Genome-Wide Analysis of a TaLEA-Introduced Transgenic Populus simonii × Populus nigra Dwarf Mutant
Abstract
:1. Introduction
2. Results and Discussion
2.1. Characterization of the Dwf1 Mutant
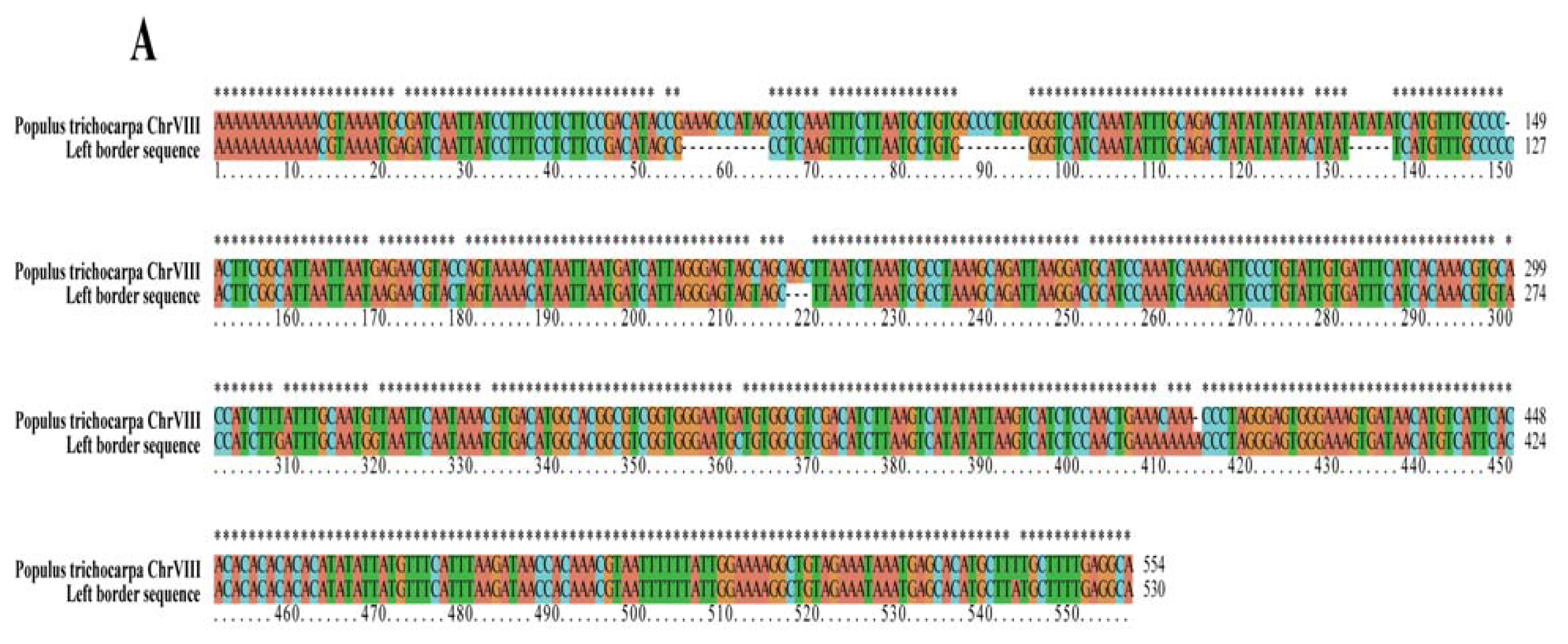
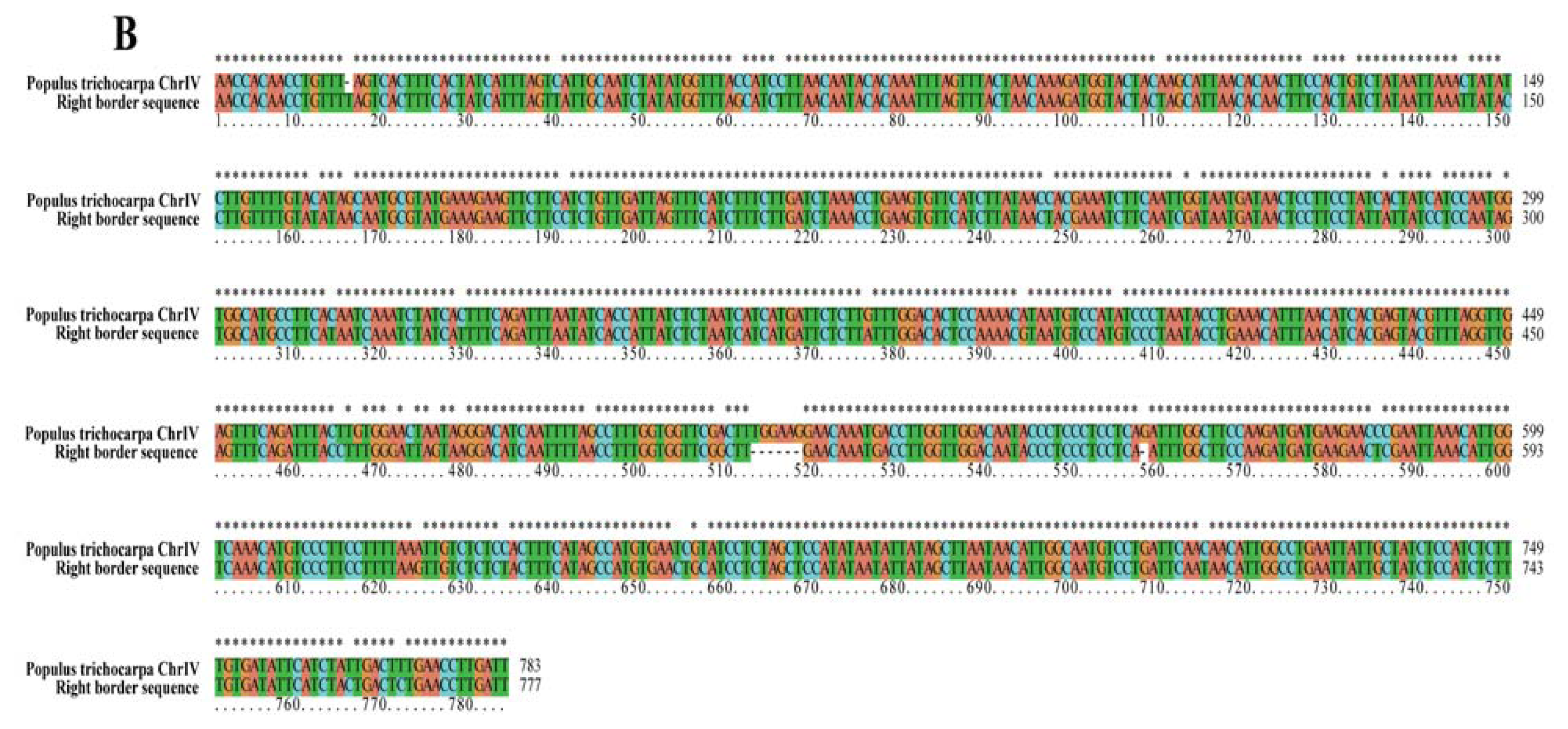
2.2. Analyses of T-DNA Flanking Sequences
2.3. Genome-Wide mRNA Expression Analysis in the Dwf1 Mutant
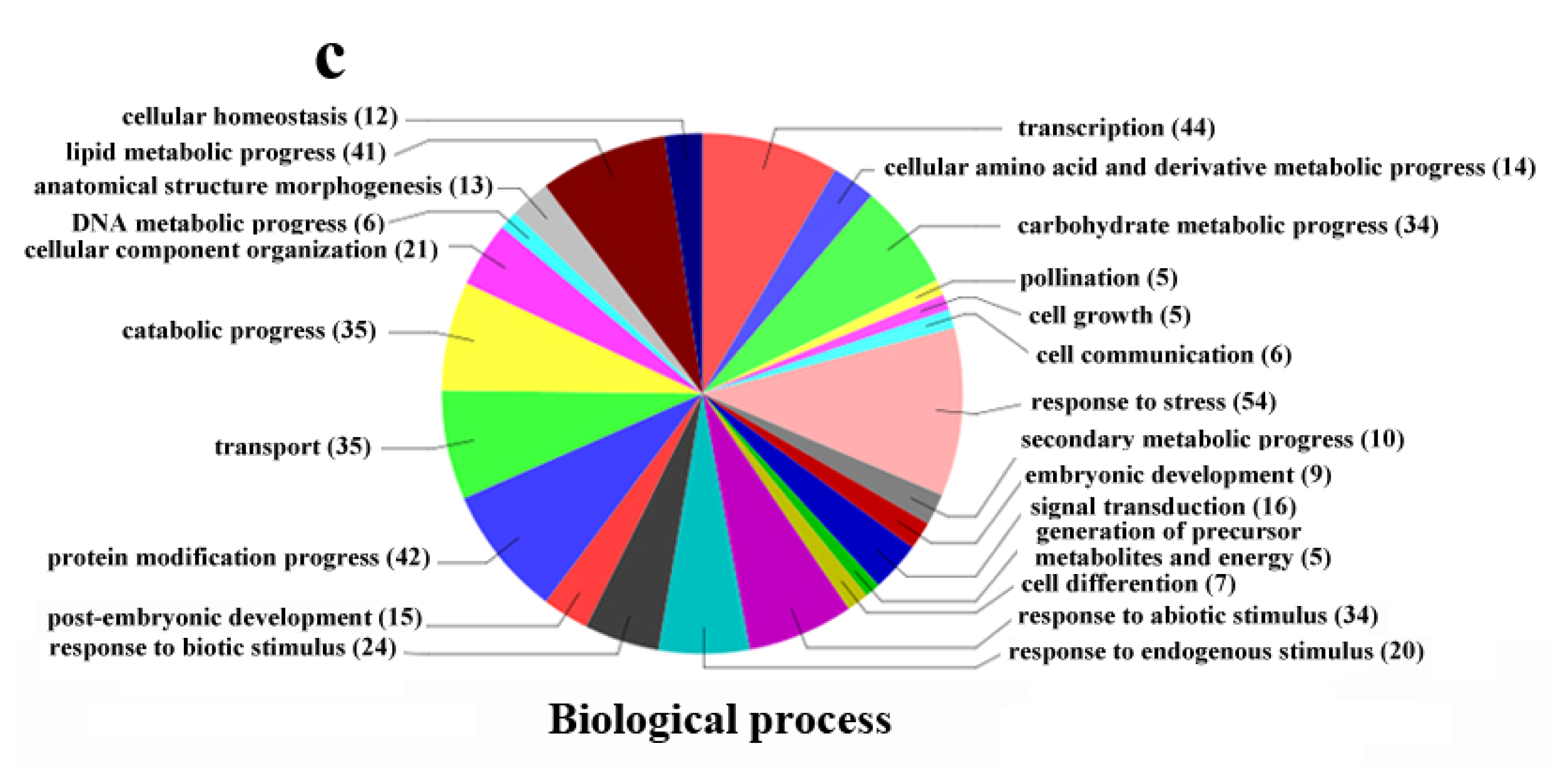
2.4. Functional Classification of Differentially Expressed Genes
2.5. Biological Pathway Analyses According to the Kyoto Encyclopedia of Genes and Genomes (KEGG)
2.6. Genes Related to Plant Hormones
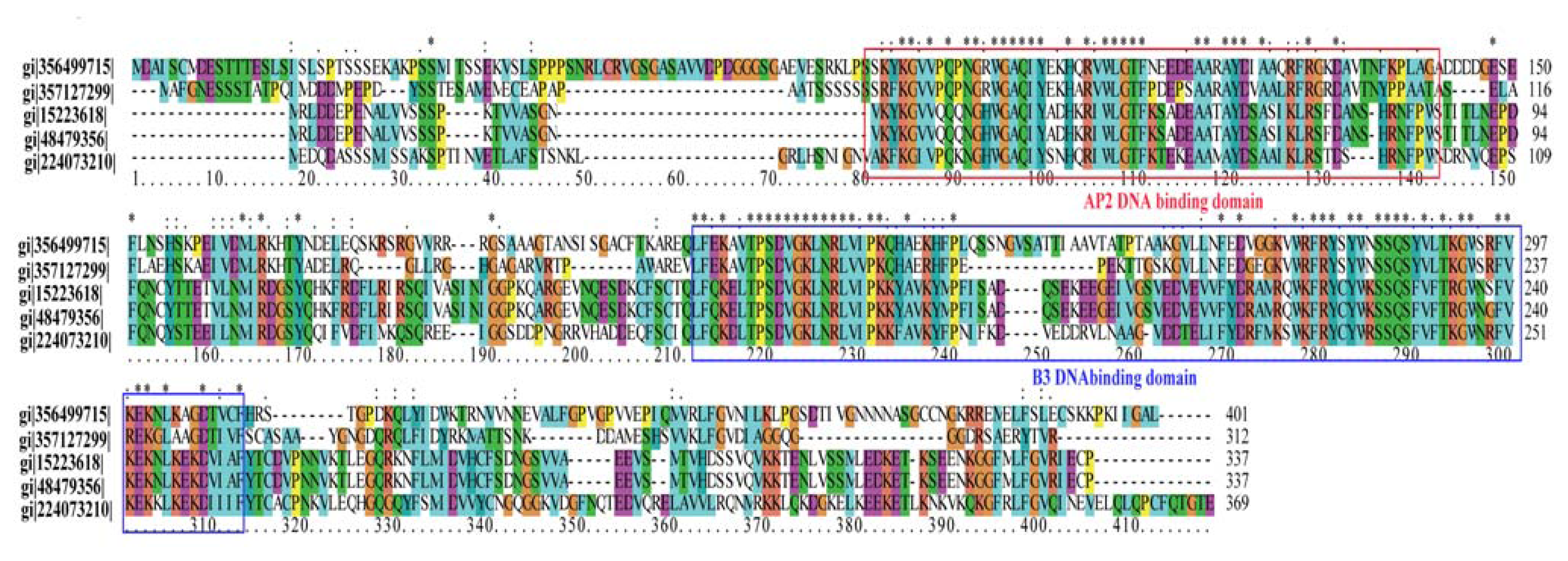
2.7. Transcription Factors with Differential Expression Patterns
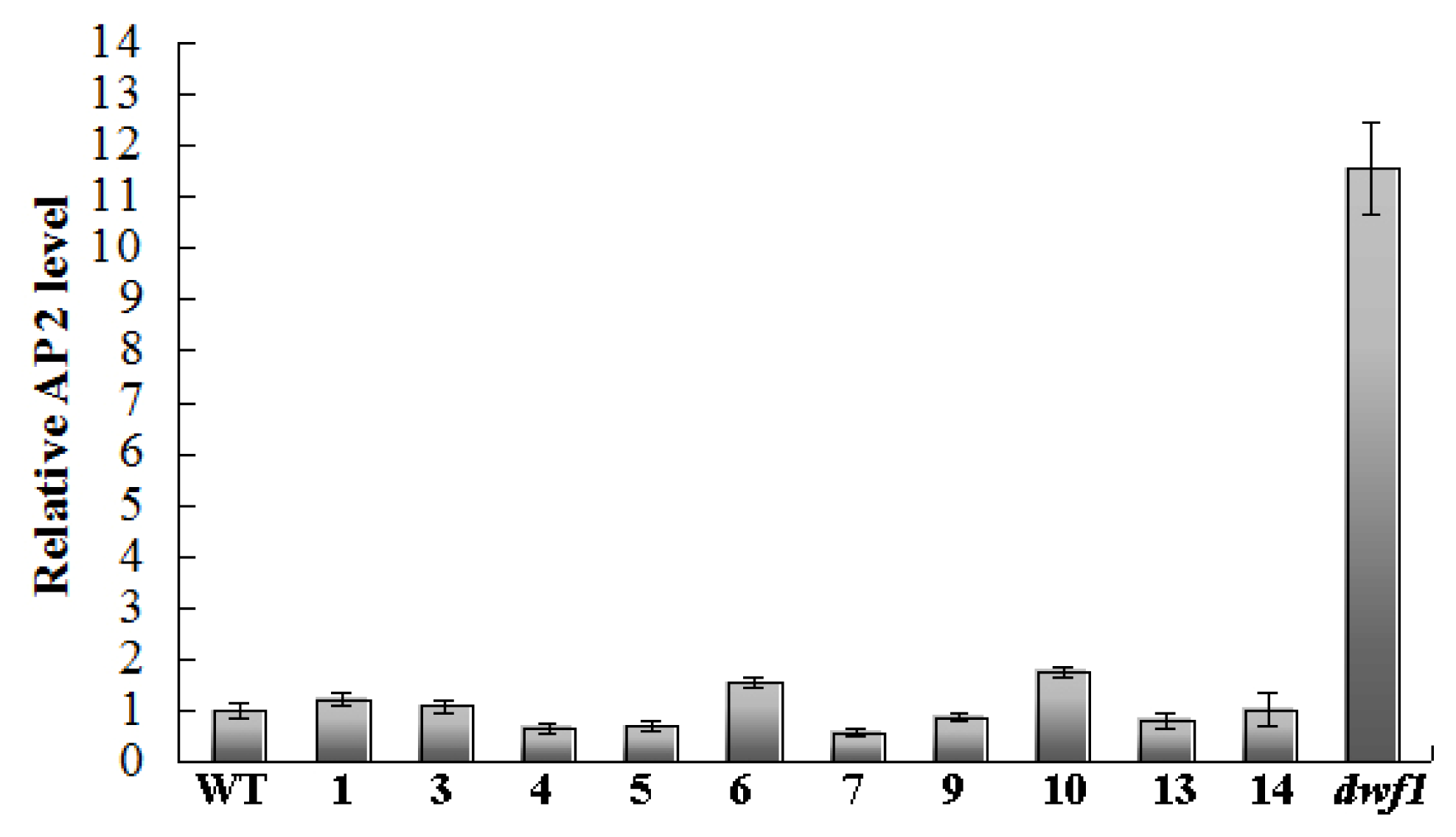
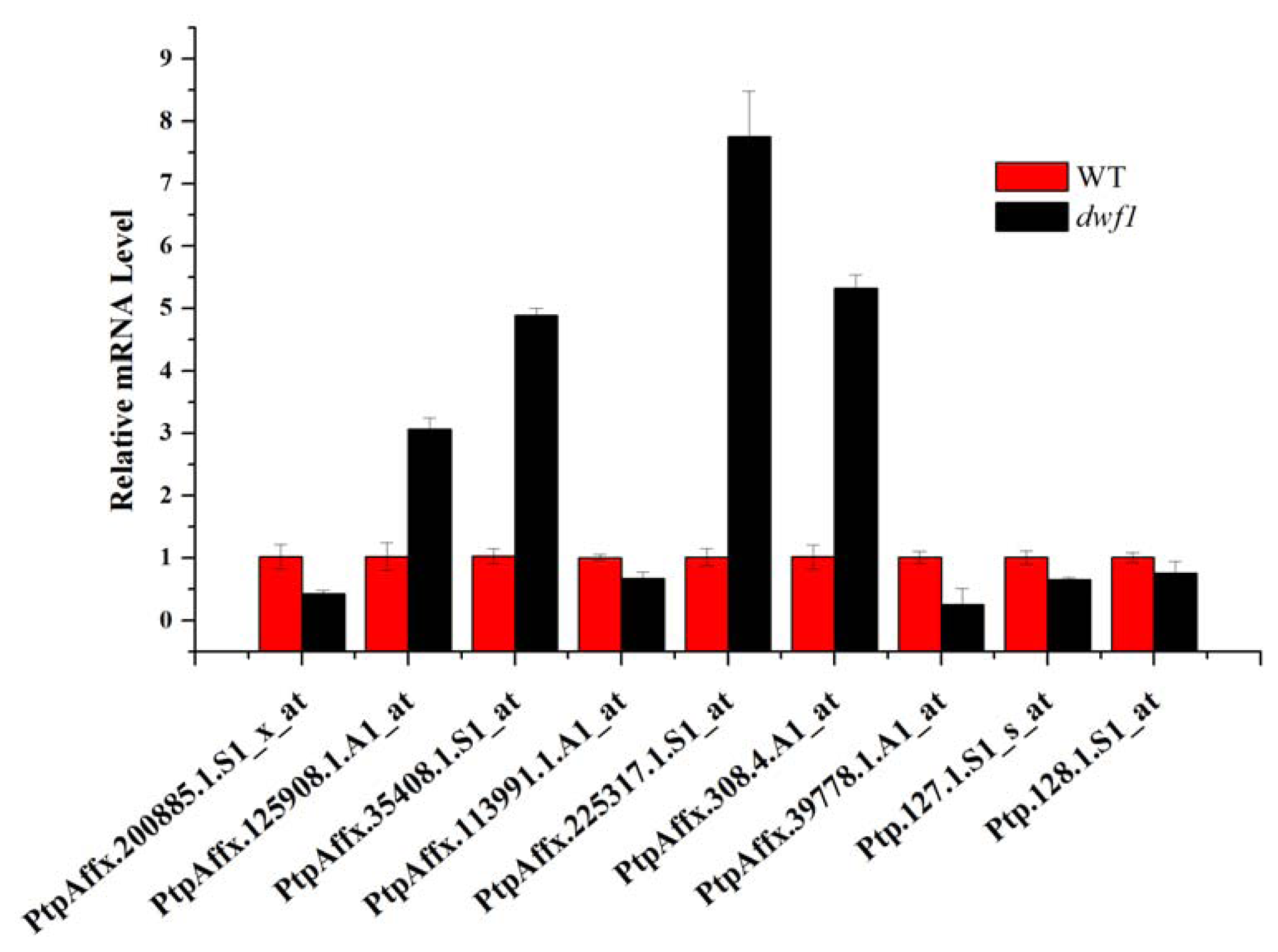
2.8. Transcript Profile Analysis of the Plant Hormones Related Genes and Transcript Factors in Other Transgenic Lines by Using Digital Gene Expression Method
3. Experimental Section
3.1. Plant Materials and Transgene
3.2. RNA Isolation and cDNA Synthesis
3.3. Quantitative Real-Time PCR
3.4. DNA Isolation and Identification of T-DNA Insertion Site
3.5. Affymetrix Microarray Analysis Experiment and Microarrays Data Analysis
3.6. DGE Analysis
3.7. Chlorophyll Content
4. Conclusions
Supplementary Information
ijms-13-02744-s001.pdfAcknowledgments
References
- Huang, H.J.; Hu, X.T.; Li, H.Y.; Jiang, J.; Li, Z.X.; Wang, L. Adverse resistance comparison of eight transgenic Populus simonii × Populus nigra Lines under NaCl Stress. For. Eng 2009, 25, 14–18. [Google Scholar]
- Liu, Y.G.; Whittier, R.F. Thermal asymmetric interlaced PCR: Automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 1995, 25, 674–681. [Google Scholar]
- Liu, Y.G.; Mitsukawa, N.; Oosumi, T.; Whittier, R.F. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 1995, 8, 457–463. [Google Scholar]
- Fujioka, S.; Yokota, T. Biosynthesis and metabolism of brassinosteroids. Annu. Rev. Plant Biol 2003, 54, 137–164. [Google Scholar]
- Magome, H.; Yamaguchi, S.; Hanada, A.; Kamiya, Y.; Oda, K. The DDF1 transcriptional activator upregulates expression of a gibberellin-deactivating gene, GA2ox7, under high-salinity stress in Arabidopsis. Plant J 2008, 56, 613–626. [Google Scholar]
- Ueguchi-Tanaka, M.; Fujisawa, Y.; Kobayashi, M.; Ashikari, M.; Iwasaki, Y.; Kitano, H.; Matsuoka, M. Rice dwarf mutant d1, which is defective in the alpha subunit of the heterotrimeric G protein, affects gibberellin signal transduction. Proc. Natl. Acad. Sci. USA 2000, 97, 11638–11643. [Google Scholar]
- Victor, B.B.; Richard, M.; David, W.P.; Caiping, M.; Stewart, B.R.; Steven, H.S. Activation tagging of a dominant gibberellin catabolism gene (GA 2-oxidase) from Poplar that regulates tree stature. Plant Physiol 2003, 132, 1283–1291. [Google Scholar]
- Hamberger, B.; Bohlmann, J. Cytochrome P450 mono-oxygenases in conifer genomes: discovery of members of the terpenoid oxygenase superfamily in spruce and pine. Biochem. Soc. Trans 2006, 34, 1209–1214. [Google Scholar]
- Ohnishi, T.; Nomura, T.; Watanabe, B.; Ohta, D.; Yokota, T.; Miyagawa, H.; Sakata, K.; Mizutani, M. Tomato cytochrome P450 CYP734A7 functions in brassinosteroid catabolism. Phytochemistry 2006, 67, 1895–1906. [Google Scholar]
- Tanabe, S.; Ashikari, M.; Fujioka, S.; Takatsuto, S.; Yoshida, S.; Yano, M.; Yoshimura, A.; Kitano, H.; Matsuoka, M.; Fujisawa, Y.; et al. A novel cytochrome P450 is implicated in brassinosteroid biosynthesis via the characterization of a rice dwarf mutant, dwarf11, with reduced seed length. Plant Cell 2005, 17, 776–790. [Google Scholar]
- Chinchilla, D.; Shan, L.; He, P.; de Vries, S.; Kemmerling, B. One for all: The receptor-associated kinase BAK1. Trends Plant Sci 2009, 14, 535–541. [Google Scholar]
- He, K.; Gou, X.; Powell, R.A.; Yang, H.; Yuan, T.; Guo, Z.; Li, J. Receptor-like protein kinases, BAK1 and BKK1, regulate a light-dependent cell-death control pathway. Plant Signal. Behav 2008, 3, 813–815. [Google Scholar]
- Kim, T.W.; Guan, S.; Sun, Y.; Deng, Z.; Tang, W.; Shang, J.X.; Burlingame, A.L.; Wang, Z.Y. Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat. Cell Biol. 2009, 11, 1254–1260. [Google Scholar]
- Yun, H.S.; Bae, Y.H.; Lee, Y.J.; Chang, S.C.; Kim, S.K.; Li, J.; Nam, K.H. Analysis of phosphorylation of the BRI1/BAK1 complex in arabidopsis reveals amino acid residues critical for receptor formation and activation of BR signaling. Mol. Cells 2009, 27, 183–190. [Google Scholar]
- NCBI. Map Viewer Home. Available online: http://www.ncbi.nlm.nih.gov/mapview/ accessed on 31 January 2007.
- Krishnaswamy, S.; Verma, S.; Rahman, M.H.; Kav, N.N. Functional characterization of four APETALA2-family genes (RAP2.6, RAP2.6L, DREB19 and DREB26) in Arabidopsis. Plant Mol. Biol. 2010, 75, 107–127. [Google Scholar]
- Licausi, F.; Giorgi, F.M.; Zenoni, S.; Osti, F.; Pezzotti, M.; Perata, P. Genomic and transcriptomic analysis of the AP2/ERF superfamily in Vitis vinifera.
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 2006, 140, 411–432. [Google Scholar]
- Nole-Wilson, S.; Krizek, B.A. DNA binding properties of the Arabidopsis floral development protein AINTEGUMENTA. Nucleic Acids Res 2000, 28, 4076–4082. [Google Scholar]
- Cui, G.H.; Feng, H.; Li, W.Y.; Wang, W.Y.; Huang, L.Q. Cloning and polymorphism analysis of SmERF in Salvia miltiorrhiza. Yao Xue Xue Bao 2011, 45, 1188–1193. [Google Scholar]
- Je, B.I.; Han, C.D. Brassinosteroid homeostasis via coordinate regulation of signaling and synthetic pathways. Plant Signal. Behav 2010, 5, 115–117. [Google Scholar]
- Je, B.I.; Piao, H.L.; Park, S.J.; Park, S.H.; Kim, C.M.; Xuan, Y.H.; Huang, J.; Do, C.Y.; An, G.; Wong, H.L.; et al. RAV-Like1 maintains brassinosteroid homeostasis via the coordinated activation of BRI1 and biosynthetic genes in rice. Plant Cell 2010, 22, 1777–1791. [Google Scholar]
- Zhao, L.; Luo, Q.; Yang, C.; Han, Y.; Li, W. A RAV-like transcription factor controls photosynthesis and senescence in soybean. Planta 2008, 227, 1389–1399. [Google Scholar]
- Woo, H.R.; Kim, J.H.; Kim, J.; Lee, U.; Song, I.J.; Lee, H.Y.; Nam, H.G.; Lim, P.O. The RAV1 transcription factor positively regulates leaf senescence in Arabidopsis. J. Exp. Bot 2010, 61, 3947–3957. [Google Scholar]
- Sharoni, A.M.; Nuruzzaman, M.; Satoh, K.; Shimizu, T.; Kondoh, H.; Sasaya, T.; Choi, I.R.; Omura, T.; Kikuchi, S. Gene structures, classification and expression models of the AP2/EREBP transcription factor family in rice. Plant Cell Physiol 2010, 52, 344–360. [Google Scholar]
- Blast2GO, version b2g_may10; BioBam Bioinformatics: Valencia, Spain, 2011.
- Ruegger, M.; Dewey, E.; Gray, W.M.; Hobbie, L.; Turner, J.; Estelle, M. The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast Grr1p. Genes Dev 1998, 12, 198–207. [Google Scholar]
- Dharmasiri, N.; Dharmasiri, S.; Estelle, M. The F-box protein TIR1 is an auxin receptor. Nature 2005, 435, 441–445. [Google Scholar]
- Dharmasiri, N.; Dharmasiri, S.; Weijers, D.; Lechner, E.; Yamada, M.; Hobbie, L.; Ehrismann, J.S.; Jurgens, G.; Estelle, M. Plant development is regulated by a family of auxin receptor F box proteins. Dev. Cell 2005, 9, 109–119. [Google Scholar]
- Thines, B.; Katsir, L.; Melotto, M.; Niu, Y.; Mandaokar, A.; Liu, G.; Nomura, K.; He, S.Y.; Howe, G.A.; Browse, J. JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature 2007, 448, 661–665. [Google Scholar]
- Craig, A.; Ewan, R.; Mesmar, J.; Gudipati, V.; Sadanandom, A. E3 ubiquitin ligases and plant innate immunity. J. Exp. Bot 2009, 60, 1123–1132. [Google Scholar]
- Maraschin, F.S.; Memelink, J.; Offringa, R. Auxin-induced, SCFTIR1-mediated poly-ubiquitination marks AUX/IAA proteins for degradation. Plant J 2009, 59, 100–109. [Google Scholar]
- Santner, A.; Calderon-Villalobos, L.; Estelle, M. Plant hormones are versatile chemical regulators of plant growth. Nat. Chem. Biol 2009, 5, 301–307. [Google Scholar]
- Yee, D.; Goring, D.R. The diversity of plant U-box E3 ubiquitin ligases: from upstream activators to downstream target substrates. J. Exp. Bot 2009, 60, 1109–1121. [Google Scholar]
- Jiang, W.; Yu, D. Arabidopsis WRKY2 transcription factor mediates seed germination and postgermination arrest of development by abscisic acid. BMC Plant Biol 2009, 9. [Google Scholar] [CrossRef]
- Koo, S.C.; Moon, B.C.; Kim, J.K.; Kim, C.Y.; Sung, S.J.; Kim, M.C.; Cho, M.J.; Cheong, Y.H. OsBWMK1 mediates SA-dependent defense responses by activating the transcription factor OsWRKY33. Biochem. Biophys. Res. Commun. 2009, 387, 365–370. [Google Scholar]
- Skinner, D.Z. Post-acclimation transcriptome adjustment is a major factor in freezing tolerance of winter wheat. Funct. Integr. Genomics 2009, 9, 513–523. [Google Scholar]
- Tao, Z.; Liu, H.; Qiu, D.; Zhou, Y.; Li, X.; Xu, C.; Wang, S. A pair of allelic WRKY genes play opposite roles in rice-bacteria interactions. Plant Physiol 2009, 151, 936–948. [Google Scholar]
- Hu, L.; Lu, H.; Liu, Q.; Chen, X.; Jiang, X. Overexpression of mtlD gene in transgenic Populus tomentosa improves salt tolerance through accumulation of mannitol. Tree Physiol 2005, 25, 1273–1281. [Google Scholar]
- Xu, M.; Zang, B.; Yao, H.S.; Huang, M.R. Isolation of high quality RNA and molecular manipulations with various tissues of Populus. Russ. J. Plant Physiol 2009, 5, 791–795. [Google Scholar]
- Porebski, S.; Bailey, L.G.; Baum, B.R. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Rep 1997, 1, 8–15. [Google Scholar]
- Allen, S.; Ellen, B.; Gernot, P.; George, A.; John, M.; David, P.; Bob, D.; Patrick, H.; Johana, B.; Cynthia, K.; et al. A high-throughput arabidopsis reverse genetics system. Plant Cell 2002, 14, 2985–2994. [Google Scholar]
- Virginia, G.T.; Robert, T.; Gilbert, C. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 2001, 98, 5116–5121. [Google Scholar]
- Joint Genome Institute. Available online: http://www.phytozome.net/poplar.php accessed on 20 August 2011.
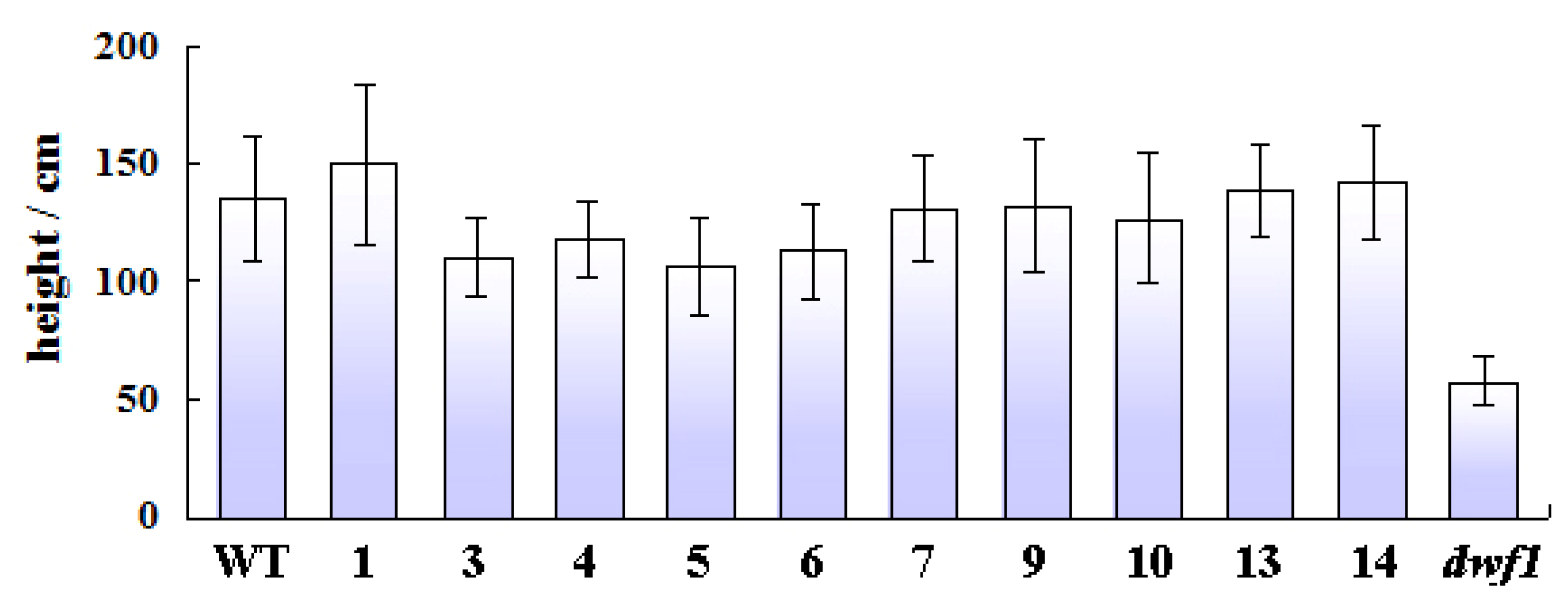
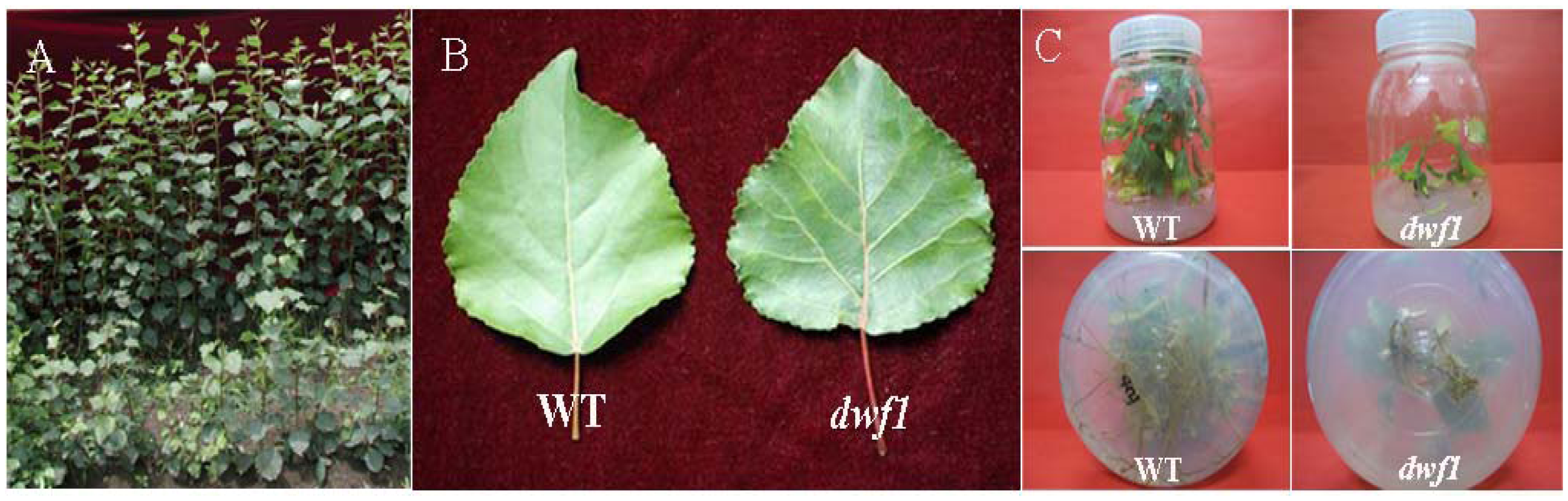


| Transgenic Line | Chlorophyll Content (SPAD) | Leaf Area (cm2) | Length/Width | Root Length (cm) |
|---|---|---|---|---|
| WT | 32.14 ± 4.04 a | 43.79 ± 13.21 a | 1.11 ± 0.14 a | 4.08 ± 0.35 a |
| XL-5 | 32.66 ± 3.71 a | 42.27 ± 9.22 a | 1.05 ± 0.13 a | 4.10 ± 0.47 a |
| XL-6 | 31.53 ± 5.74 a | 29.85 ± 5.71 b | 1.04 ± 0.13 a | 4.17 ± 0.41 a |
| dwf1 | 37.15 ± 4.24 b | 39.54 ± 10.49 a | 0.90 ± 0.08 b | 1.95 ± 0.51 b |
| Border Sequence | Start | Stop | Description | |
|---|---|---|---|---|
| Right border | T-DNA insertion sit | 10,569,359 bp | 10,570,141 bp | Chr IV |
| region displayed | 10,566,600 bp | 10,573,600 bp | no putative genes | |
| Left border | T-DNA insertion sit | 7,459,761 bp | 7,460,314 bp | Chr VIII |
| region displayed | 7,456,200 bp | 7,463,200 bp | putative AP2 transcription factor | |
| Probe Set ID | Description | Fold Change (dwf1/WT) |
|---|---|---|
| Receptors Related | ||
| PtpAffx.225074.1.S1_at | Receptor-related protein kinase-like | 13.8804 |
| PtpAffx.75698.1.A1_at | Receptor-like serine/threonine kinase | 10.9634 |
| PtpAffx.35408.1.S1_at | BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative | 7.4219 |
| PtpAffx.224953.1.S1_x_at | Leucine-rich repeat receptor-like protein kinase 1 | 5.951 |
| PtpAffx.18756.1.S1_a_at | Receptor serine/threonine kinase, putative | 5.1724 |
| Ptp.1239.2.S1_at | Vacuolar sorting receptor, putative | 4.4382 |
| PtpAffx.223006.1.S1_s_at | leucine-rich repeat receptor-like protein kinase | 4.2346 |
| PtpAffx.225656.1.S1_at | Receptor-like protein kinase homolog RK20-1 | 3.9744 |
| PtpAffx.225480.1.S1_at | (Receptor Like Protein 19); kinase/ protein binding | 3.9717 |
| PtpAffx.26522.1.S1_at | Stress-induced receptor-like kinase | 3.329 |
| PtpAffx.48409.1.S1_at | BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor | 2.8082 |
| PtpAffx.225585.1.S1_at | Receptor-like protein kinase 4, putative (RLK4) | 2.3771 |
| PtpAffx.50721.1.A1_s_at | Receptor serine-threonine protein kinase, putative | 0.3798 |
| PtpAffx.222905.1.S1_s_at | Leucine-rich repeat receptor-like protein kinase | 0.3393 |
| PtpAffx.200885.1.S1_x_at | F-box family protein | 0.4794 |
| PtpAffx.200885.1.S1_at | F-box family protein | 0.4765 |
| PtpAffx.45694.1.S1_at | F-box family protein | 0.3875 |
| PtpAffx.125908.1.A1_at | F-box protein family | 2.2595 |
| Cytochrome P450 genes | ||
| PtpAffx.225317.1.S1_at | Cytochrome P450-like | 7.2083 |
| PtpAffx.224780.1.S1_s_at | Putative cytochrome P450 | 3.1479 |
| PtpAffx.225200.1.S1_at | Cytochrome P450 family protein | 2.6336 |
| PtpAffx.124621.1.S1_s_at | Cytochrome P450 | 2.3608 |
| Ptp.2894.1.A1_at | Cytochrome P450 | 2.1906 |
| PtpAffx.203255.1.S1_s_at | Cytochrome P450, putative | 0.4878 |
| PtpAffx.220980.1.S1_s_at | Cytochrome P450 82C1-soybean | 0.4874 |
| Ptp.962.1.S1_s_at | Cytochrome P450 monooxygenase CYP77A3v2 | 0.4822 |
| PtpAffx.209025.1.S1_at | Putative cytochrome P450 | 0.4497 |
| PtpAffx.224872.1.S1_s_at | Cytochrome P450 | 0.3524 |
| PtpAffx.158051.1.A1_at | Cytochrome P450 | 0.2578 |
| Ubiquitins | ||
| PtpAffx.20179.1.A1_at | Ubiquitin | 2.3644 |
| PtpAffx.20179.3.A1_at | Ubiquitin | 2.3096 |
| PtpAffx.20179.2.S1_at | Ubiquitin | 2.0378 |
| Ptp.7320.1.S1_a_at | Ubiquitin, putative | 2.0015 |
| PtpAffx.2814.1.A1_at | Ubiquitin-protein ligase, putative | 0.4885 |
| Ptp.3476.1.S1_s_at | Ubiquitin | 0.4401 |
| PtpAffx.113991.1.A1_at | E2 ubiquitin-conjugating enzyme UBC10 | 0.4293 |
| Plant hormone response genes | ||
| PtpAffx.113518.1.S1_at | Dormancy/auxin associated protein-related | 3.2637 |
| PtpAffx.218532.1.S1_s_at | Auxin-responsive GH3 family protein | 2.3382 |
| PtpAffx.72392.1.A1_at | GH3 family protein | 2.2979 |
| Ptp.128.1.S1_at | Aux/IAA protein | 0.4928 |
| PtpAffx.123395.1.S1_at | Auxin-induced protein | 0.4888 |
| PtpAffx.204268.1.S1_at | Auxin-induced protein 15A | 0.4348 |
| PtpAffx.39778.1.A1_at | Aux/IAA protein | 0.4174 |
| Ptp.127.1.S1_s_at | Aux/IAA protein | 0.4027 |
| Probe Set ID | Description | Fold Change (dwf1/WT) |
|---|---|---|
| PtpAffx.37401.1.A1_s_at | Basic-leucine zipper (bZIP) transcription factor | 18.4804 |
| PtpAffx.107622.1.A1_at | WRKY transcription factor, putative | 17.1165 |
| PtpAffx.205219.1.S1_at | WRKY family transcription factor | 11.0952 |
| PtpAffx.211278.1.S1_at | WRKY family transcription factor | 10.6904 |
| Ptp.5995.1.S1_at | WRKY transcription factor, putative | 5.4609 |
| PtpAffx.101694.1.A1_at | WRKY transcription factor, putative | 2.6628 |
| PtpAffx.5224.1.A1_at | WRKY transcription factor 2 | 2.551 |
| PtpAffx.22176.1.A1_at | Transcriptional factor B3 | 2.4954 |
| PtpAffx.26228.1.S1_at | AP2/ERF domain-containing transcription factor | 2.4415 |
| PtpAffx.37783.1.A1_s_at | WRKY transcription factor, putative | 2.4322 |
| PtpAffx.10330.2.S1_a_at | putative zinc finger (B-box type) family protein | 2.4221 |
| PtpAffx.202579.1.S1_x_at | myb family transcription factor (MYB114) | 2.4031 |
| PtpAffx.16157.1.S1_s_at | Transcription factor GT-3b | 2.3955 |
| Ptp.4550.1.S1_at | Transcription factor LHY | 2.3908 |
| PtpAffx.132628.1.S1_s_at | Putative WRKY transcription factor 30 | 2.2339 |
| PtpAffx.34370.1.S1_at | Transcription factor, putative | 2.1851 |
| PtpAffx.221169.1.S1_at | Transcriptional factor B3 family protein | 2.1293 |
| PtpAffx.214377.1.S1_s_at | AP2-like ethylene-responsive transcription factor | 2.1219 |
| PtpAffx.219808.1.S1_at | Homeodomain transcription factor ATHB-51 | 2.1169 |
| PtpAffx.152367.1.S1_s_at | WRKY transcription factor, putative | 2.0796 |
| PtpAffx.34524.3.A1_a_at | AP2 domain-containing transcription factor | 2.0643 |
| PtpAffx.200385.1.S1_at | Ethylene-responsive transcription factor CRF2 | 2.0245 |
| PtpAffx.1802.2.S1_at | Heat shock transcription factor (HSF) | 2.0112 |
| PtpAffx.147430.1.A1_at | AP2/ERF domain-containing transcription factor | 0.4931 |
| PtpAffx.200077.1.S1_s_at | AP2-like ethylene-responsive transcription factor | 0.4527 |
| PtpAffx.68429.2.S1_at | WRKY transcription factor, putative | 0.4489 |
| Probe Set ID | Fold Change (microarrays) dwf1/WT | Fold Change (DGE) | ||
|---|---|---|---|---|
| XL-5/WT | XL-6/WT | dwf1/WT | ||
| Hormones related | ||||
| PtpAffx.35408.1.S1_at | 7.4219 | 1.02 | 1.07 | 13.81 |
| PtpAffx.223006.1.S1_s_at | 4.2346 | 1.34 | 1.76 | 3.49 |
| PtpAffx.222905.1.S1_s_at | 0.3393 | 1.34 | 1.06 | 0.66 |
| PtpAffx.200885.1.S1_x_at | 0.4794 | 1.02 | 0.98 | 0.87 |
| PtpAffx.200885.1.S1_at | 0.4765 | 0.81 | 0.78 | 0.32 |
| PtpAffx.45694.1.S1_at | 0.3875 | 1.21 | 1.34 | 0.65 |
| PtpAffx.224780.1.S1_s_at | 3.1479 | 0.82 | 1.21 | 4.51 |
| PtpAffx.225317.1.S1_at | 7.2083 | 0.91 | 0.75 | 5.55 |
| PtpAffx.20179.1.A1_at | 2.3644 | 1.71 | 1.52 | 3.12 |
| Ptp.127.1.S1_s_at | 0.4027 | 1.02 | 1.31 | 0.22 |
| PtpAffx.34524.3.A1_a_at | 2.0643 | 1.77 | 0.95 | 3.09 |
| PtpAffx.5224.1.A1_at | 2.551 | 1.55 | 1.78 | 1.87 |
| PtpAffx.200077.1.S1_s_at | 0.4527 | 1.57 | 1.88 | 0.58 |
| PtpAffx.214377.1.S1_s_at | 2.1219 | 0.53 | 0.78 | 4.59 |
© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Yuan, H.-M.; Chen, S.; Lin, L.; Wei, R.; Li, H.-Y.; Liu, G.-F.; Jiang, J. Genome-Wide Analysis of a TaLEA-Introduced Transgenic Populus simonii × Populus nigra Dwarf Mutant. Int. J. Mol. Sci. 2012, 13, 2744-2762. https://doi.org/10.3390/ijms13032744
Yuan H-M, Chen S, Lin L, Wei R, Li H-Y, Liu G-F, Jiang J. Genome-Wide Analysis of a TaLEA-Introduced Transgenic Populus simonii × Populus nigra Dwarf Mutant. International Journal of Molecular Sciences. 2012; 13(3):2744-2762. https://doi.org/10.3390/ijms13032744
Chicago/Turabian StyleYuan, Hong-Mei, Su Chen, Lin Lin, Rui Wei, Hui-Yu Li, Gui-Feng Liu, and Jing Jiang. 2012. "Genome-Wide Analysis of a TaLEA-Introduced Transgenic Populus simonii × Populus nigra Dwarf Mutant" International Journal of Molecular Sciences 13, no. 3: 2744-2762. https://doi.org/10.3390/ijms13032744
APA StyleYuan, H.-M., Chen, S., Lin, L., Wei, R., Li, H.-Y., Liu, G.-F., & Jiang, J. (2012). Genome-Wide Analysis of a TaLEA-Introduced Transgenic Populus simonii × Populus nigra Dwarf Mutant. International Journal of Molecular Sciences, 13(3), 2744-2762. https://doi.org/10.3390/ijms13032744





