MetastamiRs: Non-Coding MicroRNAs Driving Cancer Invasion and Metastasis
Abstract
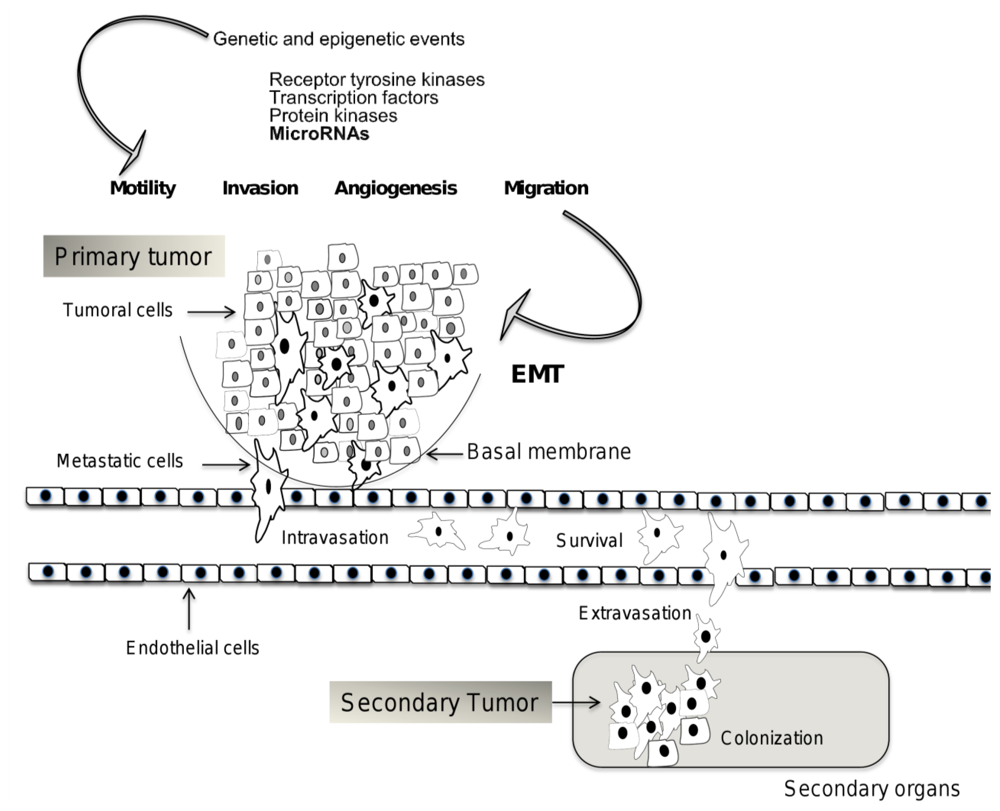
:1. Introduction
2. Mechanisms of Invasion and Metastasis
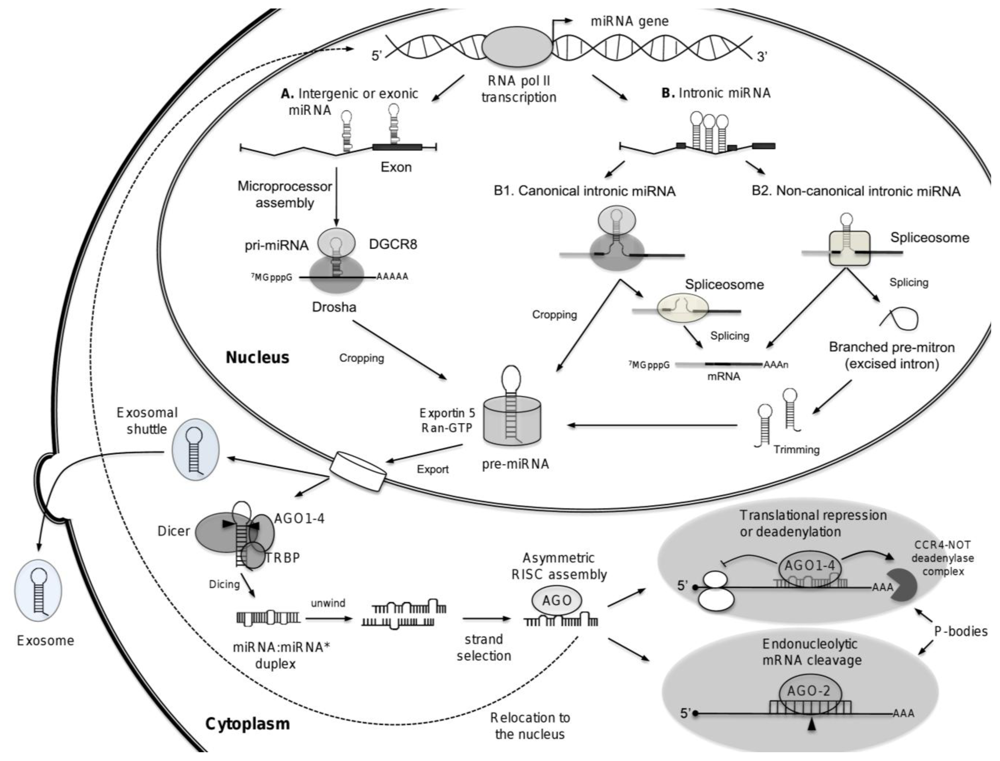
3. Biogenesis of microRNAs
4. microRNAs and Cancer: oncomiRs
5. MestastamiRs: microRNAs Regulating Metastasis
5.1. microRNAs in Breast Cancer Metastasis
5.2. MetastamiRs in Lung Cancer
5.3. MetastamiRs in Prostate Cancer
5.4. MetastamiRs in Colorectal Cancer
5.5. MetastamiRs in Gastric Cancer
5.6. MetastamiRs in Head and Neck Cancer
5.7. MetastamiRs in Cervical Carcinoma
5.8. MetastamiRs in Hepatocellular Carcinoma
6. Conclusions
Acknowledgements
Abbreviations
| miRNA | microRNA |
| RNA pol II | RNA polymerase II |
| DGCR8 | DiGeorge syndrome critical region gene 8 |
| pri-miRNA | primary-miRNA |
| pre-miRNA | precursor miRNA |
| pre-mRNA | precursor mRNA |
| 3′-UTR | 3′ untranslated region |
| RISC | RNA-induced silencing complex |
| TRBP | TAR RNA-binding protein |
| AGO | Argonaute |
References
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar]
- Lazebnik, Y. What are the hallmarks of cancer? Nat. Rev. Cancer 2010, 10, 232–233. [Google Scholar]
- Schmidt-Kittler, O.; Ragg, T.; Daskalakis, A.; Granzow, M.; Ahr, A.; Blankenstein, T.J.; Kaufmann, M.; Diebold, J.; Arnholdt, H.; Muller, P.; et al. From latent disseminated cells to overt metastasis: Genetic analysis of systemic breast cancer progression. Proc. Natl. Acad. Sci. USA 2003, 100, 7737–7742. [Google Scholar]
- Vogelstein, B.; Kinzler, K.W. The multistep nature of cancer. Trends Genet 1993, 9, 138–141. [Google Scholar]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs-microRNAs with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar]
- Nguyen, D.X.; Bos, P.D.; Massague, J. Metastasis: From dissemination to organ-specific colonization. Nat. Rev. Cancer 2009, 9, 274–284. [Google Scholar]
- Christofori, G. New signals from the invasive front. Nature 2006, 441, 444–450. [Google Scholar]
- Gupta, G.P.; Massague, J. Cancer metastasis: building a framework. Cell 2006, 127, 679–695. [Google Scholar]
- Hess, K.R.; Varadhachary, G.R.; Taylor, S.H.; Wei, W.; Raber, M.N.; Lenzi, R.; Abbruzzese, J.L. Metastatic patterns in adenocarcinoma. Cancer 2006, 106, 1624–1633. [Google Scholar]
- Fidler, I.J.; Kripke, M.L. Metastasis results from preexisting variant cells within a malignant tumor. Science 1977, 197, 893–895. [Google Scholar]
- Liotta, L.A.; Kohn, E.C. The microenvironment of the tumour-host interface. Nature 2001, 411, 375–379. [Google Scholar]
- Friedl, P.; Wolf, K. Tumour-cell invasion and migration: diversity and escape mechanisms. Nat. Rev. Cancer 2003, 3, 362–374. [Google Scholar]
- Stein, U.; Walther, W.; Arlt, F.; Schwabe, H.; Smith, J.; Fichtner, I.; Birchmeier, W.; Schlag, P.M. MACC1, a newly identified key regulator of HGF-MET signaling, predicts colon cancer metastasis. Nat. Med 2009, 15, 59–67. [Google Scholar]
- Guo, C.; Sah, J.F.; Beard, L.; Willson, J.K.; Markowitz, S.D.; Guda, K. The noncoding RNA, miR-126, suppresses the growth of neoplastic cells by targeting phosphatidylinositol 3-kinase signaling and is frequently lost in colon cancers. Genes Chromosomes Cancer 2008, 47, 939–946. [Google Scholar]
- Ramaswamy, S.; Ross, K.N.; Lander, E.S.; Golub, T.R. A molecular signature of metastasis in primary solid tumors. Nat. Genet 2003, 33, 49–54. [Google Scholar]
- Chambers, A.F.; Groom, A.C.; MacDonald, I.C. Dissemination and growth of cancer cells in metastatic sites. Nat. Rev. Cancer 2002, 2, 563–572. [Google Scholar]
- Hynes, R.O. Integrins: Bidirectional, allosteric signaling machines. Cell 2002, 110, 673–687. [Google Scholar]
- Biancone, L.; Araki, M.; Araki, K.; Vassalli, P.; Stamenkovic, I. Redirection of tumor metastasis by expression of E-selectin in vivo. J. Exp. Med 1996, 183, 581–587. [Google Scholar]
- Yang, J.; Weinberg, R.A. Epithelial-mesenchymal transition: At the crossroads of development and tumor metastasis. Dev. Cell 2008, 14, 818–829. [Google Scholar]
- Hotary, K.; Li, X.Y.; Allen, E.; Stevens, S.L.; Weiss, S.J. A cancer cell metalloprotease triad regulates the basement membrane transmigration program. Genes Dev 2006, 20, 2673–2686. [Google Scholar]
- Hiraoka, N.; Allen, E.; Apel, I.J.; Gyetko, M.R.; Weiss, S.J. Matrix metalloproteinases regulate neovascularization by acting as pericellular fibrinolysins. Cell 1998, 95, 365–377. [Google Scholar]
- Folkman, J. Role of angiogenesis in tumor growth and metastasis. Semin. Oncol 2002, 29, 15–18. [Google Scholar]
- Yin, J.J.; Selander, K.; Chirgwin, J.M.; Dallas, M.; Grubbs, B.G.; Wieser, R.; Massagué, J.; Mundy, G.R.; Guise, T.A. TGF-β signaling blockade inhibits PTHrP secretion by breast cancer cells and bone metastases development. J. Clin. Invest 1999, 103, 197–206. [Google Scholar]
- Kang, Y.; Siegel, P.M.; Shu, W.; Drobnjak, M.; Kakonen, S.M.; Cordón-Cardo, C.; Guise, T.A.; Massagué, J.A. Multigenic program mediating breast cancer metastasis to bone. Cancer Cell 2003, 3, 537–549. [Google Scholar]
- Mundy, G.R. Metastasis to bone: Causes, consequences and therapeutic opportunities. Nat. Rev. Cancer 2002, 2, 584–593. [Google Scholar]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar]
- Kim, V.N. MicroRNA biogenesis: Coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol 2005, 6, 376–385. [Google Scholar]
- Ruby, J.G.; Jan, C.H.; Bartel, D.P. Intronic microRNA precursors that bypass Drosha processing. Nature 2007, 448, 83–86. [Google Scholar]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J 2004, 23, 4051–4060. [Google Scholar]
- Cai, X.; Hagedorn, C.H.; Cullen, B.R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 2004, 10, 1957–1966. [Google Scholar]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Radmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar]
- Lund, E.; Guttinger, S.; Calado, A.; Dahlberg, J.E.; Kutay, U. Nuclear export of microRNA precursors. Science 2004, 303, 95–98. [Google Scholar]
- Yi, R.; Qin, Y.; Macara, I.G.; Cullen, B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev 2003, 17, 3011–3016. [Google Scholar]
- Vasudevan, S.; Tong, Y.; Steitz, J.A. Switching from repression to activation: microRNAs can up-regulate translation. Science 2007, 318, 1931–1934. [Google Scholar]
- Hwang, H.W.; Wentzel, E.A.; Mendell, J.T. A hexanucleotide element directs microRNA nuclear import. Science 2007, 315, 97–100. [Google Scholar]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol 2007, 9, 654–659. [Google Scholar]
- Zhang, B.; Pan, X.; Cobb, G.P.; Anderson, T.A. microRNAs as oncogenes and tumor suppressors. Dev. Biol 2007, 302, 1–12. [Google Scholar]
- Hernando, E. microRNAs and cancer: Role in tumorigenesis, patient classification and therapy. Clin. Transl. Oncol 2007, 9, 155–160. [Google Scholar]
- Calin, G.A.; Sevignani, C.; Dumitru, C.D.; Hyslop, T.; Noch, E.; Yendamuri, S.; Shimizu, M.; Rattan, S.; Bullrich, F.; Negrini, M.; et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. USA 2004, 101, 2999–3004. [Google Scholar]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar]
- Karube, Y.; Tanaka, H.; Osada, H.; Tomida, S.; Tatematsu, Y.; Yanagisawa, K.; Yatabe, Y.; Takamizawa, J.; Miyoshi, S.; Mitsudomi, T.; et al. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci 2005, 96, 111–115. [Google Scholar]
- Merritt, W.M.; Lin, Y.G.; Han, L.Y.; Kamat, A.A.; Spannuth, W.A.; Schmandt, R.; Urbauer, D.; Pennacchio, L.A.; Cheng, J.-F.; Nick, A.M.; et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. N. Engl. J. Med 2008, 359, 2641–2650. [Google Scholar]
- Carmell, M.A.; Xuan, Z.; Zhang, M.Q.; Hannon, G.J. The Argonaute family: Tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev 2002, 16, 2733–2742. [Google Scholar]
- Nelson, P.; Kiriakidou, M.; Sharma, A.; Maniataki, E.; Mourelatos, Z. The microRNA world: Small is mighty. Trends Biochem. Sci 2003, 28, 534–540. [Google Scholar]
- Steeg, P.S. Tumor metastasis: Mechanistic insights and clinical challenges. Nat. Med 2006, 12, 895–904. [Google Scholar]
- Hurst, D.R.; Edmonds, M.D.; Welch, D.R. Metastamir: The field of metastasis-regulatory microRNA is spreading. Cancer Res 2009, 69, 7495–749. [Google Scholar]
- Ma, L.; Teruya-Feldstein, J.; Weinberg, R.A. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 2007, 449, 682–688. [Google Scholar]
- Huang, Q.; Gumireddy, K.; Schrier, M.; le Sage, C.; Nagel, R.; Nair, S.; Egan, D.A.; Li, A.; Huang, G.; Klein-Szanto, A.J.; et al. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat. Cell Biol 2008, 10, 292–210. [Google Scholar]
- Tavazoie, S.F.; Alarcon, C.; Oskarsson, T.; Padua, D.; Wang, Q.; Bos, P.D.; Gerald, W.L.; Massague, J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature 2008, 451, 147–152. [Google Scholar]
- Yang, J.; Mani, S.A.; Donaher, J.L.; Ramaswamy, S.; Itzykson, R.A.; Come, C.; Savagner, P.; Gitelman, I.; Richardson, A.; Weinberg, R.A. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 2004, 117, 927–939. [Google Scholar]
- Botas, J. Control of morphogenesis and differentiation by HOM/Hox genes. Curr. Opin. Cell Biol 1993, 5, 1015–1022. [Google Scholar]
- Makiyama, K.; Hamada, J.; Takada, M.; Murakawa, K.; Takahashi, Y.; Tada, M.; Tamoto, E.; Shindo, G.; Matsunaga, A.; Teramoto, K.; et al. Aberrant expression of HOX genes in human invasive breast carcinoma. Oncol. Rep 2005, 13, 673–679. [Google Scholar]
- Ma, L.; Reinhardt, F.; Pan, E.; Soutschek, J.; Bhat, B.; Marcusson, E.G.; Teruya-Feldstein, J.; Bell, G.W.; Weinberg, R.A. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat. Biotechnol 2010, 28, 341–347. [Google Scholar]
- Li, F.; Tiede, B.; Massagué, J.; Kang, Y. Beyond tumorigenesis: Cancer stem cells in metastasis. Cell Res 2007, 17, 3–14. [Google Scholar]
- Voorhoeve, P.M.; le Sage, C.; Schrier, M.; Gillis, A.J.; Stoop, H.; Nagel, R.; Liu, Y.P.; van Duijse, J.; Drost, J.; Griekspoor, A.; et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell 2006, 124, 1169–1181. [Google Scholar]
- Oskarsson, T.; Acharyya, S.; Zhang, X.H.; Vanharanta, S.; Tavazoie, S.F.; Morris, P.G.; Downey, R.J.; Manova-Todorova, K.; Brogi, E.; Massagué, J. Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nat. Med 2011, 17, 867–874. [Google Scholar]
- van de Wetering, M.; Oosterwegel, M.; van Norren, K.; Clevers, H. Sox-4, an Sry-like HMG box protein, is a transcriptional activator in lymphocytes. EMBO J 1993, 12, 3847–3854. [Google Scholar]
- Png, K.J.; Halberg, N.; Yoshida, M.; Tavazoie, S.F. A microRNA regulon that mediates endothelial recruitment and metastasis by cancer cells. Nature 2011. [Google Scholar] [CrossRef]
- Bhaumik, D.; Scott, G.K.; Schokrpur, S.; Patil, C.K.; Campisi, J.; Benz, C.C. Expression of microRNA-146 suppresses NF-kappaB activity with reduction of metastatic potential in breast cancer cells. Oncogene 2008, 27, 5643–5647. [Google Scholar]
- Lin, S.L.; Chiang, A.; Chang, D.; Ying, S.Y. Loss of mir-146a function in hormone-refractory prostate cancer. RNA 2008, 14, 417–424. [Google Scholar]
- Hurst, D.R.; Edmonds, M.D.; Scott, G.K.; Benz, C.C.; Vaidya, K.S.; Welch, D.R. Breast cancer metastasis suppressor 1 up-regulates miR-146, which suppresses breast cancer metastasis. Cancer Res 2009, 69, 1279–1283. [Google Scholar]
- Valastyan, S.; Reinhardt, F.; Benaich, N.; Calogrias, D.; Szász, A.M.; Wang, Z.C.; Brock, J.E.; Richardson, A.L.; Weinberg, R.A. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell 2009, 137, 1032–1046. [Google Scholar]
- Li, X.F.; Yan, P.J.; Shao, Z.M. Downregulation of miR-193b contributes to enhance urokinase-type plasminogen activator (uPA) expression and tumor progression and invasion in human breast cancer. Oncogene 2009, 28, 3937–3948. [Google Scholar]
- Lowery, A.J.; Miller, N.; Dwyer, R.M.; Kerin, M.J. Dysregulated miR-183 inhibits migration in breast cancer cells. BMC Cancer 2010, 10, 502. [Google Scholar]
- Sachdeva, M.; Mo, Y.Y. MicroRNA-145 suppresses cell invasion and metastasis by directly targeting mucin 1. Cancer Res 2010, 70, 378–387. [Google Scholar]
- Yu, Z.; Willmarth, N.E.; Zhou, J.; Katiyar, S.; Wang, M.; Liu, Y.; McCue, P.A.; Quong, A.A.; Lisanti, M.P.; Pestell, R.G. MicroRNA 17/20 inhibits cellular invasion and tumor metastasis in breast cancer by heterotypic signaling. Proc. Natl. Acad. Sci. USA 2010, 107, 8231–8236. [Google Scholar]
- Ma, L.; Young, J.; Prabhala, H.; Pan, E.; Mestdagh, P.; Muth, D.; Teruya-Feldstein, J.; Reinhardt, F.; Onder, T.T.; Valastyan, S.; et al. MiR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat. Cell Biol 2010, 12, 247–256. [Google Scholar]
- Dykxhoorn, D.M.; Wu, Y.; Xie, H.; Yu, F.; Lal, A.; Petrocca, F.; Martinvalet, D.; Song, E.; Lim, B.; Lieberman, J. MiR-200 enhances mouse breast cancer cell colonization to form distant metastases. PLoS One 2009, 4, e7181. [Google Scholar]
- Vetter, G.; Saumet, A.; Moes, M.; Vallar, L.; le Béchec, A.; Laurini, C.; Sabbah, M.; Arar, K.; Theillet, C.; Lecellier, C.H.; et al. MiR-661 expression in SNAI1-induced epithelial to mesenchymal transition contributes to breast cancer cell invasion by targeting Nectin-1 and StarD10 messengers. Oncogene 2010, 29, 4436–4448. [Google Scholar]
- Li, Y.; Zhang, M.; Chen, H.; Dong, Z.; Ganapathy, V.; Thangaraju, M.; Huang, S. Ratio of miR-196s to HOXC8 messenger RNA correlates with breast cancer cell migration and metastasis. Cancer Res 2010, 70, 7894–7904. [Google Scholar]
- Jemal, A.; Siegel, R.; Xu, J.; Ward, E. Cancer statistics 2010. CA Cancer J. Clin 2010, 60, 277–300. [Google Scholar]
- Le, X.F.; Merchant, O.; Bast, R.C.; Calin, G.A. The roles of microRNAs in the cancer invasion-metastasis cascade. Cancer Microenviron 2010, 3, 137–147. [Google Scholar]
- Wang, Z.; Longo, P.A.; Tarrant, M.K.; Kim, K.; Head, S.; Leahy, D.J.; Cole, P.A. Mechanistic insights into the activation of oncogenic forms of EGF receptor. Nat. Struct. Mol. Biol 2011, in press. [Google Scholar]
- Wang, G.; Mao, W.; Zheng, S.; Ye, J. Epidermal growth factor receptor-regulated miR-125a-5p-a metastatic inhibitor of lung cancer. FEBS J 2009, 276, 5571–5578. [Google Scholar]
- Yu, S.L.; Chen, H.Y.; Chang, G.C.; Chen, C.Y.; Chen, H.W.; Singh, S.; Cheng, C.L.; Yu, C.J.; Lee, Y.C.; Chen, H.S.; et al. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell 2008, 13, 48–57. [Google Scholar]
- Jiang, L.; Huang, Q.; Zhang, S.; Zhang, Q.; Chang, J.; Qiu, X.; Wang, E. Hsa-miR-125a-3p and hsa-miR-125a-5p are downregulated in non-small cell lung cancer and have inverse effects on invasion and migration of lung cancer cells. BMC Cancer 2010, 10, 318–330. [Google Scholar]
- Wang, G.; Mao, W.; Zheng, S. MicroRNA-183 regulates Ezrin expression in lung cancer cells. FEBS Lett 2008, 582, 3663–3668. [Google Scholar]
- Sarver, A.L.; Li, L.; Subramanian, S. MicroRNA miR-183 functions as an oncogene by targeting the transcription factor EGR1 and promoting tumor cell migration. Cancer Res 2010, 70, 9570–9580. [Google Scholar]
- Zhu, W.; Liu, X.; He, J.; Chen, D.; Hunag, Y.; Zhang, Y.K. Overexpression of members of the microRNA-183 family is a risk factor for lung cancer: A case control study. BMC Cancer 2011, 11, 393–399. [Google Scholar]
- Yanaihara, N.; Caplen, N.; Bowman, E.; Seike, M.; Kumamoto, K.; Yi, M.; Stephens, R.M.; Okamoto, A.; Yokota, J.; Tanaka, T.; et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 2006, 9, 189–198. [Google Scholar]
- Crawford, M.; Brawner, E.; Batte, K.; Yu, L.; Hunter, M.G.; Otterson, G.A.; Nuovo, G.; Marsh, C.B.; Nana-Sinkam, S.P. MicroRNA-126 inhibits invasion in non-small cell lung carcinoma cell lines. Biochem. Biophys. Res. Commun 2008, 373, 607–612. [Google Scholar]
- Korpal, M.; Kang, Y. The emerging role of miR-200 family of microRNAs in epithelial-mesenchymal transition and cancer metastasis. RNA Biol 2008, 5, 115–119. [Google Scholar]
- Roybal, J.D.; Zang, Y.; Ahn, Y.H.; Yang, Y.; Gibbons, D.L.; Baird, B.N.; Alvarez, C.; Thilaganathan, N.; Liu, D.D.; Saintigny, P.; et al. MiR-200 inhibits lung adenocarcinoma cell invasion and metastasis by targeting Flt1/VEGFR1. Mol. Cancer Res 2011, 9, 25–35. [Google Scholar]
- Garofalo, M.; di Leva, G.; Romano, G.; Nuovo, G.; Suh, S.S.; Ngankeu, A.; Taccioli, C.; Pichiorri, F.; Alder, H.; Secchiero, P.; et al. MiR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell 2009, 16, 498–509. [Google Scholar]
- Shen, J.; Todd, N.W.; Zhang, H.; Yu, L.; Lingxiao, X.; Mei, Y.; Guarnera, M.; Liao, J.; Chou, A.; Lu, C.L.; et al. Plasma microRNAs as potential biomarkers for non-small-cell lung cancer. Lab. Invest 2011, 91, 579–587. [Google Scholar]
- Zhu, S.; Wu, H.; Wu, F.; Nie, D.; Sheng, S.; Mo, Y.Y. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res 2008, 18, 350–359. [Google Scholar]
- Nicoloso, M.S.; Spizzo, R.; Shimizu, M.; Rossi, S.; Calin, G.A. MicroRNAs-the micro steering wheel of tumour metastases. Nat. Rev. Cancer 2009, 9, 293–302. [Google Scholar]
- Wang, R.; Wang, Z.X.; Yang, J.S.; Pan, X.; De, W.; Chen, L.B. MicroRNA-451 functions as a tumor suppressor in human non-small cell lung cancer by targeting ras-related protein 14 (RAB14). Oncogene 2011, 30, 2644–2658. [Google Scholar]
- Lujambio, A.; Calin, G.A.; Villanueva, A.; Ropero, S.; Sánchez-Céspedes, M.; Blanco, D.; Montuenga, L.M.; Rossi, S.; Nicoloso, M.S.; Faller, W.J.; et al. microRNA DNA methylation signature for human cancer metastasis. Proc. Natl. Acad. Sci. USA 2008, 105, 13556–13561. [Google Scholar]
- Baffa, R.; Fassan, M.; Volinia, S.; O’Hara, B.; Liu, C.G.; Palazzo, J.P.; Gardiman, M.; Rugge, M.; Gomella, L.G.; Croce, C.M.; et al. MicroRNA expression profiling of human metastatic cancers identifies cancer gene targets. J. Pathol 2009, 219, 214–221. [Google Scholar]
- Arora, S.; Ranade, A.R.; Tran, N.L.; Nasser, S.; Sridhar, S.; Korn, R.L.; Ross, J.T.; Dhruv, H.; Foss, K.M.; Sibenaller, Z.; et al. MicroRNA-328 is associated with (non-small) cell lung cancer (NSCLC) brain metastasis and mediates NSCLC migration. Int. J. Cancer 2011, in press. [Google Scholar]
- Shi, X.B.; Tepper, C.G.; White, R.W.; Krichevsky, A.M.; Gabriely, G. MicroRNAs and prostate cancer. J. Cell. Mol. Med 2008, 5A, 1456–1465. [Google Scholar]
- Volinia, S.; Calin, G.A.; Liu, C.G.; Ambs, S.; Cimmino, A.; Petrocca, F.; Visone, R.; Iorio, M.; Roldo, C.; Ferracin, M.; et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 2006, 103, 2257–2261. [Google Scholar]
- Li, T.; Li, D.; Sha, J.; Sun, P.; Huang, Y. MicroRNA-21 directly targets MARCKS and promotes apoptosis resistance and invasion in prostate cancer cells. Biochem. Biophys. Res. Commun 2009, 383, 280–285. [Google Scholar]
- Watahiki, A.; Wang, Y.; Morris, J.; Dennis, K.; O’Dwyer, H.M.; Gleave, M.; Gout, P.W.; Wang, Y. MicroRNAs associated with metastatic prostate cancer. PLoS One 2011, in press. [Google Scholar]
- Wu, H.; Zhu, S.; Mo, Y.Y. Suppression of cell growth and invasion by miR-205 in breast cancer. Cell Res 2009, 19, 439–448. [Google Scholar]
- Gandellini, P.; Folini, M.; Longoni, N.; Pennati, M.; Binda, M.; Colecchia, M.; Salvioni, R.; Supino, R.; Moretti, R.; Limonta, P.; et al. MiR-205 exerts tumor-suppressive functions in human prostate through down-regulation of protein kinase C-epsilon. Cancer Res 2009, 69, 2287–2295. [Google Scholar]
- Cao, Q.; Yu, J.; Dhanasekaran, S.M.; Kim, J.H.; Mani, R.S.; Tomlins, S.A.; Mehra, R.; Laxman, B.; Cao, X.; Kleer, C.G.; et al. Repression of E-cadherin by the polycomb group protein EZH2 in cancer. Oncogene 2008, 27, 7274–7284. [Google Scholar]
- Varambally, S.; Cao, Q.; Mani, R.S.; Shankar, S.; Wang, X.; Ateeq, B.; Laxman, B.; Cao, X.; Jing, X.; Ramnarayanan, K.; et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science 2008, 322, 1695–1699. [Google Scholar]
- Peng, X.; Guo, W.; Liu, T.; Wang, X.; Tu, X.; Xiong, D.; Chen, S.; Lai, Y.; Du, H.; Chen, G.; et al. Identification of miRs-143 and -145 that is associated with bone metastasis of prostate cancer and involved in the regulation of EMT. PLoS One 2011, in press. [Google Scholar]
- Yang, L.; Belaguli, N.; Berger, D.H. MicroRNA and colorectal cancer. World J. Surg 2009, 33, 638–646. [Google Scholar]
- Schetter, A.J.; Harris, C.C. Alterations of microRNAs contribute to colon carcinognesis. Semin. Oncol 2011, 38, 734–742. [Google Scholar]
- Gabriely, G.; Wurdinger, T.; Kesari, S.; Esau, C.C.; Burchard, J.; Linsley, P.S.; Krichevsky, A.M. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol. Cell. Biol 2008, 28, 5369–5380. [Google Scholar]
- Takayama, T.; Miyanishi, K.; Hayashi, T.; Sato, Y.; Niitsu, Y. Colorectal cancer: Genetics of development and metastasis. J. Gastroenterol 2006, 41, 185–192. [Google Scholar]
- Dews, M.; Homayouni, A.; Yu, D.; Murphy, D.; Sevignani, C.; Wentzel, E.; Furth, E.F.; Lee, W.M.; Enders, G.H.; Mendell, J.T.; et al. Augmentation of tumor angiogenesis by a MYC-activated microRNA cluster. Nat. Genet 2006, 38, 1060–1065. [Google Scholar]
- Guo, J.; Miao, Y.; Xiao, B.; Huan, R.; Jiang, Z.; Meng, D.; Wang, Y. Differential expression of microRNA species in human gastric cancer versus non-tumorous tissues. J. Gastroenterol. Hepatol 2009, 24, 652–657. [Google Scholar]
- Brenner, B.; Hoshen, M.B.; Purim, O.; David, M.B.; Ashkenazi, K.; Marshak, G.; Kundel, Y.; Brenner, R.; Morgenstern, S.; Halpern, M.; et al. MicroRNAs as a potential prognostic factor in gastric cancer. World J. Gastroenterol 2011, 17, 3976–3985. [Google Scholar]
- Zhang, Z.; Liu, S.; Shi, R.; Zhao, G. miR-27 promotes human gastric cancer cell metastasis by inducing epithelial-to-mesenchymal transition. Cancer Genet 2011, 204, 486–491. [Google Scholar]
- Liang, S.; He, L.; Zhao, X.; Miao, Y.; Gu, Y.; Guo, C.; Xue, Z.; Dou, W.; Hu, F.; Wu, K.; et al. MicroRNA let-7f inhibits tumor invasion and metastasis by targeting MYH9 in human gastric cancer. PLoS One 2011, 6, e18409. [Google Scholar]
- Zheng, B.; Liang, L.; Wang, C.; Huang, S.; Cao, X.; Zha, R.; Liu, L.; Jia, D.; Tian, Q.; Wu, J.; et al. MicroRNA-148a suppresses tumor cell invasion and metastasis by downregulating ROCK1 in gastric cancer. Clin. Cancer Res 2011, in press. [Google Scholar]
- Guo, X.B.; Jing, C.Q.; Li, L.P.; Zhang, L.; Shi, Y.L.; Wang, J.S.; Liu, J.L.; Li, C.S. Down-regulation of miR-622 in gastric cancer promotes cellular invasion and tumor metastasis by targeting ING1 gene. World J. Gastroenterol 2011, 17, 1895–1902. [Google Scholar]
- Feng, L.; Xie, Y.; Zhang, H.; Wu, Y. miR-107 targets cyclin-dependent kinase 6 expression, induces cell cycle G1 arrest and inhibits invasion in gastric cancer cells. Med. Oncol 2011, in press. [Google Scholar]
- Li, X.; Zhang, Y.; Shi, Y.; Dong, G.; Liang, J.; Han, Y.; Wang, X.; Zhao, Q.; Ding, J.; Wu, K.; et al. MicroRNA-107, an oncogene microRNA that regulates tumour invasion and metastasis by targeting DICER1 in gastric cancer. J. Cell. Mol. Med 2011. [Google Scholar] [CrossRef]
- Zhang, C.; Han, L.; Zhang, A.; Fu, Y.; Yue, X.; Wang, G.; Jia, Z.; Pu, P.; Zhang, Q.; Kang, C. MicroRNA-221 and microRNA-222 regulate gastric carcinoma cell proliferation and radioresistance by targeting PTEN. BMC Cancer 2010, 10, 367. [Google Scholar]
- Wang, H.J.; Ruan, H.J.; He, X.J.; Ma, Y.Y.; Jiang, X.T.; Xia, Y.J.; Ye, Z.Y.; Tao, H.Q. MicroRNA-101 is downregulated in gastric cancer and involved in cell migration and invasion. Eur. J. Cancer 2010, 46, 2295–2303. [Google Scholar]
- Wu, W.Y.; Xue, X.Y.; Chen, Z.J.; Han, S.L.; Huang, Y.P.; Zhang, L.F.; Zhu, G.B.; Shen, X. Potentially predictive microRNAs of gastric cancer with metastasis to lymph node. World J. Gastroenterol 2011, 17, 3645–3651. [Google Scholar]
- Liu, X.; Jiang, L.; Wang, A.; Yu, J.; Shi, F.; Zhou, X. MicroRNA-138 suppresses invasion and promotes apoptosis in head and neck squamous cell carcinoma cell lines. Cancer Lett 2009, 286, 217–222. [Google Scholar]
- Wong, T.S.; Liu, X.B.; Wong, B.Y.; Ng, R.W.; Yuen, A.P.; Wei, W.I. Mature miR-184 as potential oncogenic microRNA of squamous cell carcinoma of tongue. Clin. Cancer Res 2008, 14, 2588–2592. [Google Scholar]
- Mitomo, S.; Maesawa, C.; Ogasawara, S.; Iwaya, T.; Shibazaki, M.; Yashima-Abo, A.; Kotani, K.; Oikawa, H.; Sakurai, E.; Izutsu, N.; et al. Downregulation of miR-138 is associated with overexpression of human telomerase reverse transcriptase protein in human anaplastic thyroid carcinoma cell lines. Cancer Sci 2008, 99, 280–286. [Google Scholar]
- Piccinin, S.; Gasparotto, D.; Vukosavljevic, T.; Barzan, L.; Sulfaro, S.; Maestro, R.; Boiocchi, M. Microsatellite instability in squamous cell carcinomas of the head and neck related to field cancerization phenomena. Br. J. Cancer 1998, 78, 1147–1151. [Google Scholar]
- Hogg, R.P.; Honorio, S.; Martinez, A.; Agathanggelou, A.; Dallol, A.; Fullwood, P.; Weichselbaum, R.; Kuo, M.J.; Maher, E.R.; Latif, F. Frequent 3p allele loss and epigenetic inactivation of the RASSF1A tumour suppressor gene from region 3p21.3 in head and neck squamous cell carcinoma. Eur. J. Cancer 2002, 38, 1585–1592. [Google Scholar]
- Wang, X.; Gleich, L.; Pavelic, Z.P.; Li, Y.Q.; Gale, N.; Hunt, S.; Gluckman, J.L.; Stambrook, P.J. Cervical metastases of head and neck squamous cell carcinoma correlate with loss of heterozygosity on chromosome 16q. Int. J. Oncol 1999, 14, 557–561. [Google Scholar]
- Liu, X.; Yu, J.; Jiang, L.; Wang, A.; Shi, F.; Ye, H.; Zhou, X. MicroRNA-222 regulates cell invasion by targeting matrix metalloproteinase 1 (MMP1) and manganese superoxide dismutase 2 (SOD2) in tongue squamous cell carcinoma cell lines. Cancer Genomics Proteomics 2009, 6, 131–139. [Google Scholar]
- Hu, L.; Ibrahim, S.; Liu, C.; Skaar, J.; Pagano, M.; Karpatkin, S. Thrombin induces tumor cell cycle activation and spontaneous growth by down-regulation of p27Kip1, in association with the up-regulation of Skp2 and MiR-222. Cancer Res 2009, 69, 3374–3381. [Google Scholar]
- Miller, T.E.; Ghoshal, K.; Ramaswamy, B.; Roy, S.; Datta, J.; Shapiro, C.L.; Jacob, S.; Majumder, S. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J. Biol. Chem 2008, 83, 29897–29903. [Google Scholar]
- Nelson, K.K.; Melendez, J.A. Mitochondrial redox control of matrix metalloproteinases. Free Radic. Biol. Med 2004, 37, 768–784. [Google Scholar]
- Nelson, K.K.; Ranganathan, A.C.; Mansouri, J.; Rodriguez, A.M.; Providence, K.M.; Rutter, J.L.; Pumiglia, K.; Bennett, J.A.; Melendez, J.A. Elevated sod2 activity augments matrix metalloproteinase expression: Evidence for the involvement of endogenous hydrogen peroxide in regulating metastasis. Clin. Cancer Res 2003, 9, 424–432. [Google Scholar]
- Yang, J.Q.; Zhao, W.; Duan, H.; Robbins, M.E.; Buettner, G.R.; Oberley, L.W.; Domann, F.E. v-Ha-RaS oncogene upregulates the 92-kDa type IV collagenase (MMP-9) gene by increasing cellular superoxide production and activating NF-kappaB. Free Radic. Biol. Med 2001, 31, 520–529. [Google Scholar]
- Sengupta, S.; den Boon, J.A.; Chen, I.H.; Newton, M.A.; Stanhope, S.A.; Cheng, Y.J.; Chen, C.J.; Hildesheim, A.; Sugden, B.; Ahlquist, P. MicroRNA 29c is down-regulated in nasopharyngeal carcinomas, up-regulating mRNAs encoding extracellular matrix proteins. Proc. Natl. Acad. Sci. USA 2008, 105, 5874–5878. [Google Scholar]
- Kaufman, L.J.; Brangwynne, C.P.; Kasza, K.E.; Filippidi, E.; Gordon, V.D.; Deisboeck, T.S.; Weitz, D.A. Glioma expansion in collagen I matrices: Analyzing collagen concentration-dependent growth and motility patterns. Biophys. J 2005, 89, 635–650. [Google Scholar]
- Chintala, S.K.; Sawaya, R.; Gokaslan, Z.L.; Rao, J.S. The effect of type III collagen on migration and invasion of human glioblastoma cell lines in vitro. Cancer Lett 1996, 102, 57–63. [Google Scholar]
- Kuratomi, Y.; Nomizu, M.; Tanaka, K.; Ponce, M.L.; Komiyama, S.; Kleinman, H.K.; Yamada, Y. Laminin gamma 1 chain peptide, C-16 (KAFDITYVRLKF), promotes migration, MMP-9 secretion, and pulmonary metastasis of B16–F10 mouse melanoma cells. Br. J. Cancer 2002, 86, 1169–1173. [Google Scholar]
- Song, S.Y.; Nomizu, M.; Yamada, Y.; Kleinman, H.K. Liver metastasis formation by laminin-1 peptide (LQVQLSIR)-adhesion selected B16–F10 melanoma cells. Int. J. Cancer 1997, 71, 436–441. [Google Scholar]
- Shintani, Y.; Hollingsworth, M.A.; Wheelock, M.J.; Johnson, K.R. Collagen I promotes metastasis in pancreatic cancer by activating c-Jun NH2-terminal kinase 1 and up-regulating N-cadherin expression. Cancer Res 2006, 66, 11745–11753. [Google Scholar]
- Sengupta, S.; den Boon, J.A.; Chen, I.H.; Newton, M.A.; Dahl, D.B.; Chen, M.; Cheng, Y.J.; Westra, W.H.; Chen, C.J.; Hildesheim, A.; et al. Genome-wide expression profiling reveals EBV-associated inhibition of MHC class I expression in nasopharyngeal carcinoma. Cancer Res 2006, 66, 7999–8006. [Google Scholar]
- Song, L.B.; Liao, W.T.; Mai, H.Q.; Zhang, H.Z.; Zhang, L.; Li, M.Z.; Hou, JH.; Fu, L.W.; Huang, W.L.; Zeng, Y.X.; et al. The clinical significance of twist expression in nasopharyngeal carcinoma. Cancer Lett. 2006, 242, 258–265. [Google Scholar]
- Horikawa, T.; Yang, J.; Kondo, S.; Yoshizaki, T.; Joab, I.; Furukawa, M.; Pagano, J.S. Twist and epithelial-mesenchymal transition are induced by the EBV oncoprotein latent membrane protein 1 and are associated with metastatic nasopharyngeal carcinoma. Cancer Res 2007, 67, 1970–1978. [Google Scholar]
- Li, G.; Wu, Z.; Peng, Y.; Liu, X.; Lu, J.; Wang, L.; Pan, Q.; He, M.L.; Li, X.P. MicroRNA-10b induced by Epstein-Barr virus-encoded latent membrane protein-1 promotes the metastasis of human nasopharyngeal carcinoma cells. Cancer Lett 2010, 299, 29–36. [Google Scholar]
- Su, X.; Chakravarti, D.; Cho, M.S.; Liu, L.; Gi, Y.J.; Lin, Y.L.; Leung, M.L.; El-Naggar, A.; Creighton, C.J.; Suraokar, M.B.; et al. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature 2010, 467, 986–990. [Google Scholar]
- Yang, A.; Kaghad, M.; Wang, Y.; Gillett, E.; Fleming, M.D.; Dötsch, V.; Andrews, N.C.; Caput, D.; McKeon, F. p63, A p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol. Cell 1998, 2, 305–316. [Google Scholar]
- Kumar, M.S.; Pester, R.E.; Chen, C.Y.; Lane, K.; Chin, C.; Lu, J.; Kirsch, D.G.; Golub, T.R.; Jacks, T. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev 2009, 23, 2700–2704. [Google Scholar]
- He, L.; He, X.; Lim, L.P.; de Stanchina, E.; Xuan, Z.; Liang, Y.; Xue, W.; Zender, L.; Magnus, J.; Ridzon, D.; et al. A microRNA component of the p53 tumour suppressor network. Nature 2007, 447, 1130–1134. [Google Scholar]
- Lee, Y.; Yang, X.; Huang, Y.; Fan, H.; Zhang, Q.; Wu, Y.; Li, J.; Hasina, R.; Cheng, C.; Lingen, M.W.; et al. Network modeling identifies molecular functions targeted by miR-204 to suppress head and neck tumor metastasis. PLoS Comput. Biol 2010, 6, e1000730. [Google Scholar]
- Abou-Elhamd, K.E.; Habib, T.N.; Moussa, A.E.; Badawy, B.S. The role of genetic susceptibility in head and neck squamous cell carcinoma. Eur. Arch. Otorhinolaryngol 2008, 265, 217–222. [Google Scholar]
- Chung, C.H.; Parker, J.S.; Karaca, G.; Wu, J.; Funkhouser, W.K.; Moore, D.; Butterfoss, D.; Xiang, D.; Zanation, A.; Yin, X.; et al. Molecular classification of head and neck squamous cell carcinomas using patterns of gene expression. Cancer Cell 2004, 5, 489–500. [Google Scholar]
- Parkin, D.M.; Bray, F.; Ferlay, J.; Pisani, P. Estimating the world cancer burden: Globocan 2000. Int. J. Cancer 2001, 94, 153–156. [Google Scholar]
- Bosch, F.X.; Manos, M.M.; Munoz, N.; Sherman, M.; Jansen, A.M.; Peto, J.; Schiffman, M.H.; Moreno, V.; Kurman, R.; Shah, K.V. Prevalence of human papillomavirus in cervical cancer: A worldwide perspective. International biological study on cervical cancer (IBSCC) Study Group. J. Natl. Cancer Inst 1995, 87, 796–802. [Google Scholar]
- Scheffner, M.; Werness, B.A.; Huibregtse, J.M.; Levine, A.J.; Howley, P.M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 1990, 63, 1129–1136. [Google Scholar]
- Boyer, S.N.; Wazer, D.E.; Band, V. E7 protein of human papilloma virus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res 1996, 56, 4620–4624. [Google Scholar]
- Munger, K.; Basile, J.R.; Duensing, S.; Eichten, A.; Gonzalez, S.L.; Grace, M.; Zacny, V.L. Biological activities and molecular targets of the human papillomavirus E7 oncoprotein. Oncogene 2001, 20, 7888–7898. [Google Scholar]
- Rapp, L.; Chen, J.J. The papillomavirus E6 proteins. Biochim. Biophys. Acta 1998, 1378, F1–F19. [Google Scholar]
- Antinore, M.J.; Birrer, M.J.; Patel, D.; Nader, L.; McCance, D.J. The human papillomavirus type 16 E7 gene product interacts with and trans-activates the AP1 family of transcription factors. EMBO J 1996, 15, 1950–1960. [Google Scholar]
- Prathapam, T.; Kuhne, C.; Banks, L. The HPV-16 E7 oncoprotein binds Skip and suppresses its transcriptional activity. Oncogene 2001, 20, 7677–7685. [Google Scholar]
- Duensing, S.; Munger, K. The human papillomavirus type 16 E6 and E7 oncoproteins independently induce numerical and structural chromosome instability. Cancer Res 2002, 62, 7075–7082. [Google Scholar]
- Aguda, B.D.; Kim, Y.; Piper-Hunter, M.G.; Friedman, A.; Marsh, C.B. MicroRNA regulation of a cancer network: consequences of the feedback loops involving miR-17-92, E2F, and MYC. Proc. Natl. Acad. Sci. USA 2008, 105, 19678–19683. [Google Scholar]
- Jevnaker, A.M.; Khuu, C.; Kjøle, E.; Bryne, M.; Osmundsen, H. Expression of members of the miRNA17-92 cluster during development and in carcinogenesis. J. Cell. Physiol 2011, 226, 2257–2266. [Google Scholar]
- Hong, L.; Lai, M.; Chen, M.; Xie, C.; Liao, R.; Kang, Y.J.; Xiao, C.; Hu, W.Y.; Han, J.; Sun, P. The miR-17-92 cluster of microRNAs confers tumorigenicity by inhibiting oncogene-induced senescence. Cancer Res 2010, 70, 8547–8557. [Google Scholar]
- Zheng, Z.M.; Wang, X. Regulation of cellular miRNA expression by human papillomaviruses. Biochim. Biophys. Acta 2011, 1809, 668–677. [Google Scholar]
- Qiang, R.; Wang, F.; Shi, L.Y.; Liu, M.; Chen, S.; Wan, H.Y.; Li, Y.X.; Li, X.; Gao, S.Y.; Sun, B.C.; et al. Plexin-B1 is a target of miR-214 in cervical cancer and promotes the growth and invasion of HeLa cells. Int. J. Biochem. Cell Biol 2011, 43, 632–641. [Google Scholar]
- Hu, X.; Schwarz, J.K.; Lewis, J.S., Jr; Huettner, P.C.; Rader, J.S.; Deasy, J.O.; Grigsby, P.W.; Wang, X. A microRNA expression signature for cervical cancer prognosis. Cancer Res. 2010, 70, 1441–1448. [Google Scholar]
- Lee, J.W.; Choi, C.H.; Choi, J.J.; Park, Y.A.; Kim, S.J.; Hwang, S.Y.; Kim, W.Y.; Kim, T.J.; Lee, J.H.; Kim, B.G.; et al. Altered microRNA expression in cervical carcinomas. Clin. Cancer Res 2008, 14, 2535–2542. [Google Scholar]
- Lupberger, J.; Hildt, E. Hepatitis B virus-induced oncogenesis. World J. Gastroenterol 2007, 13, 74–81. [Google Scholar]
- Farazi, P.A.; DePinho, R.A. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat. Rev. Cancer 2006, 6, 674–687. [Google Scholar]
- El-Serag, H.B.; Rudolph, K.L. Hepatocellular carcinoma: Epidemiology and molecular carcinogenesis. Gastroenterology 2007, 132, 2557–2576. [Google Scholar]
- Pilkis, S.J.; Granner, D.K. Molecular physiology of the regulation of hepatic gluconeogenesis and glycolysis. Annu. Rev. Physiol 1992, 54, 885–909. [Google Scholar]
- Yu, D.Y.; Moon, H.B.; Son, J.K.; Jeong, S.; Yu, S.L.; Yoon, H.; Han, Y.M.; Lee, C.S.; Park, J.S.; Lee, C.H.; et al. Incidence of hepatocellular carcinoma in transgenic mice expressing the hepatitis B virus X-protein. J. Hepatol 1999, 31, 123–132. [Google Scholar]
- Su, F.; Theodosis, C.N.; Schneider, R.J. Role of NF-kappaB and MYC proteins in apoptosis induced by hepatitis B virus HBx protein. J. Virol 2001, 75, 215–225. [Google Scholar]
- Coulouarn, C.; Factor, V.M.; Andersen, J.B.; Durkin, M.E.; Thorgeirsson, S.S. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene 2009, 28, 3526–3536. [Google Scholar]
- Kota, J.; Chivukula, R.R.; O’Donnell, K.A.; Wentzel, E.A.; Montgomery, C.L.; Hwang, H.W.; Chang, T.C.; Vivekanandan, P.; Torbenson, M.; Clark, K.R.; et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell 2009, 137, 1005–1017. [Google Scholar]
- Gramantieri, L.; Ferracin, M.; Fornari, F.; Veronese, A.; Sabbioni, S.; Liu, C.G.; Calin, G.A.; Giovannini, C.; Ferrazzi, E.; Grazi, G.L.; et al. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res 2007, 67, 6092–6099. [Google Scholar]
- Xu, T.; Zhu, Y.; Xiong, Y.; Ge, Y.Y.; Yun, J.P.; Zhuang, S.M. MicroRNA-195 suppresses tumorigenicity and regulates G1/S transition of human hepatocellular carcinoma cells. Hepatology 2009, 50, 113–121. [Google Scholar]
- Meng, F.; Henson, R.; Wehbe-Janek, H.; Ghoshal, K.; Jacob, S.T.; Patel, T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 2007, 133, 647–658. [Google Scholar]
- Wong, Q.W.; Ching, A.K.; Chan, A.W.; Choy, K.W.; To, K.F.; Lai, P.B.; Wong, N. MiR-222 overexpression confers cell migratory advantages in hepatocellular carcinoma through enhancing AKT signaling. Clin. Cancer Res 2010, 16, 867–875. [Google Scholar]
- Wang, B.; Hsu, S.H.; Majumder, S.; Kutay, H.; Huang, W.; Jacob, S.T.; Ghoshal, K. TGFbeta-mediated upregulation of hepatic miR-181b promotes hepatocarcinogenesis by targeting TIMP3. Oncogene 2010, 29, 1787–1797. [Google Scholar]
- Zhang, X.; Liu, S.; Hu, T.; Liu, S.; He, Y.; Sun, S. Up-regulated microRNA-143 transcribed by nuclear factor kappa B enhances hepatocarcinoma metastasis by repressing fibronectin expression. Hepatology 2009, 50, 490–499. [Google Scholar]
- Yang, F.; Yin, Y.; Wang, F.; Wang, Y.; Zhang, L.; Tang, Y.; Sun, S. MiR-17-5p Promotes migration of human hepatocellular carcinoma cells through the p38 mitogen-activated protein kinase-heat shock protein 27 pathway. Hepatology 2010, 51, 1614–1623. [Google Scholar]

© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Lopez-Camarillo, C.; Marchat, L.A.; Arechaga-Ocampo, E.; Perez-Plasencia, C.; Moral-Hernandez, O.d.; Castaneda-Ortiz, E.J.; Rodriguez-Cuevas, S. MetastamiRs: Non-Coding MicroRNAs Driving Cancer Invasion and Metastasis. Int. J. Mol. Sci. 2012, 13, 1347-1379. https://doi.org/10.3390/ijms13021347
Lopez-Camarillo C, Marchat LA, Arechaga-Ocampo E, Perez-Plasencia C, Moral-Hernandez Od, Castaneda-Ortiz EJ, Rodriguez-Cuevas S. MetastamiRs: Non-Coding MicroRNAs Driving Cancer Invasion and Metastasis. International Journal of Molecular Sciences. 2012; 13(2):1347-1379. https://doi.org/10.3390/ijms13021347
Chicago/Turabian StyleLopez-Camarillo, Cesar, Laurence A. Marchat, Elena Arechaga-Ocampo, Carlos Perez-Plasencia, Oscar del Moral-Hernandez, Elizabeth J. Castaneda-Ortiz, and Sergio Rodriguez-Cuevas. 2012. "MetastamiRs: Non-Coding MicroRNAs Driving Cancer Invasion and Metastasis" International Journal of Molecular Sciences 13, no. 2: 1347-1379. https://doi.org/10.3390/ijms13021347
APA StyleLopez-Camarillo, C., Marchat, L. A., Arechaga-Ocampo, E., Perez-Plasencia, C., Moral-Hernandez, O. d., Castaneda-Ortiz, E. J., & Rodriguez-Cuevas, S. (2012). MetastamiRs: Non-Coding MicroRNAs Driving Cancer Invasion and Metastasis. International Journal of Molecular Sciences, 13(2), 1347-1379. https://doi.org/10.3390/ijms13021347




