Molecular Mechanisms of RADA16-1 Peptide on Fast Stop Bleeding in Rat Models
Abstract
:1. Introduction
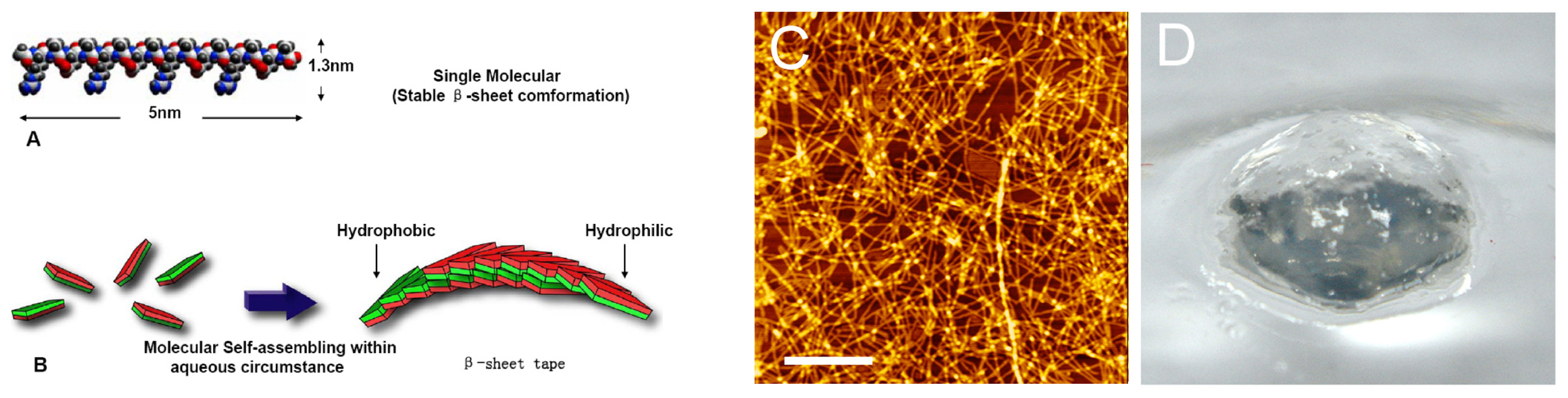
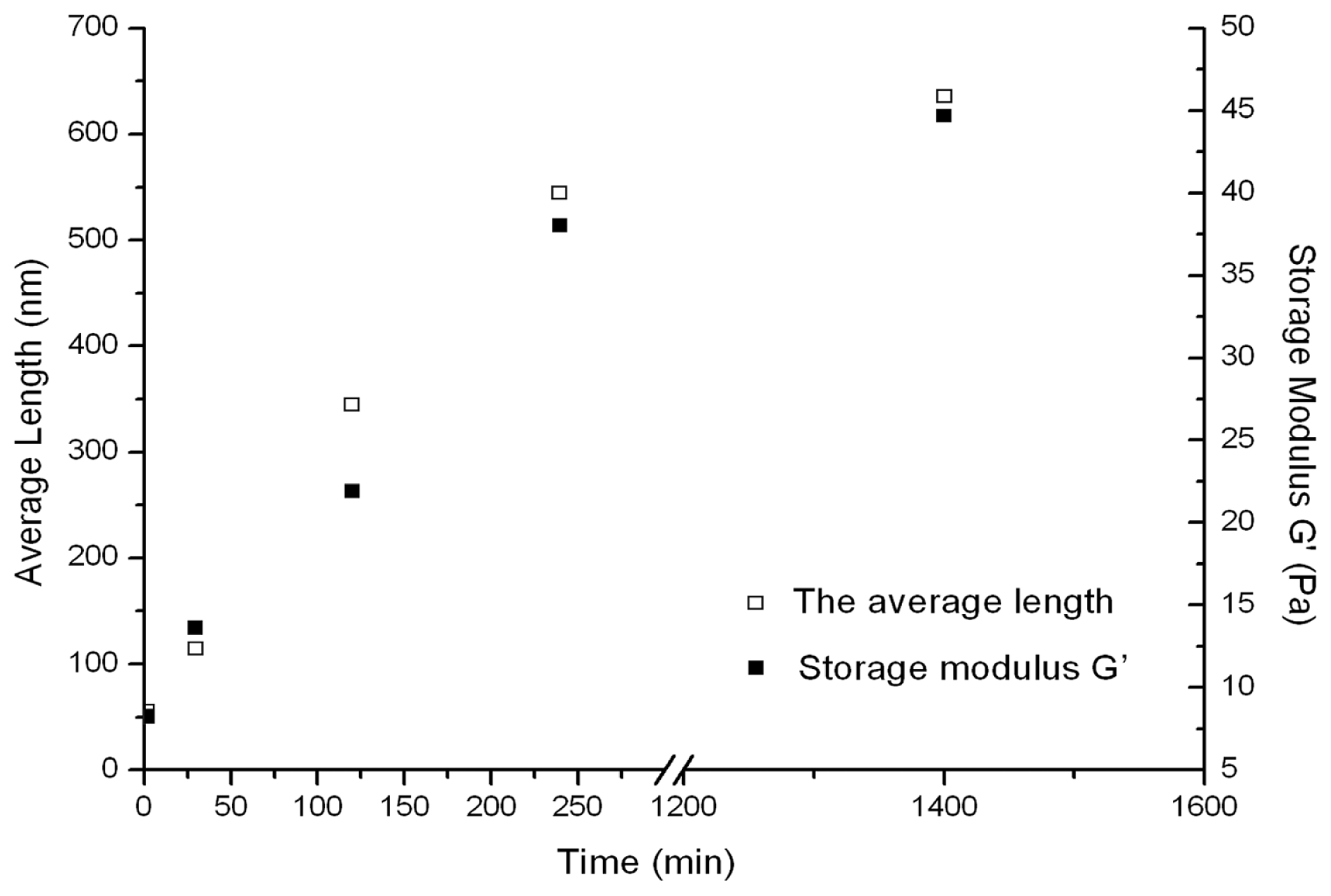
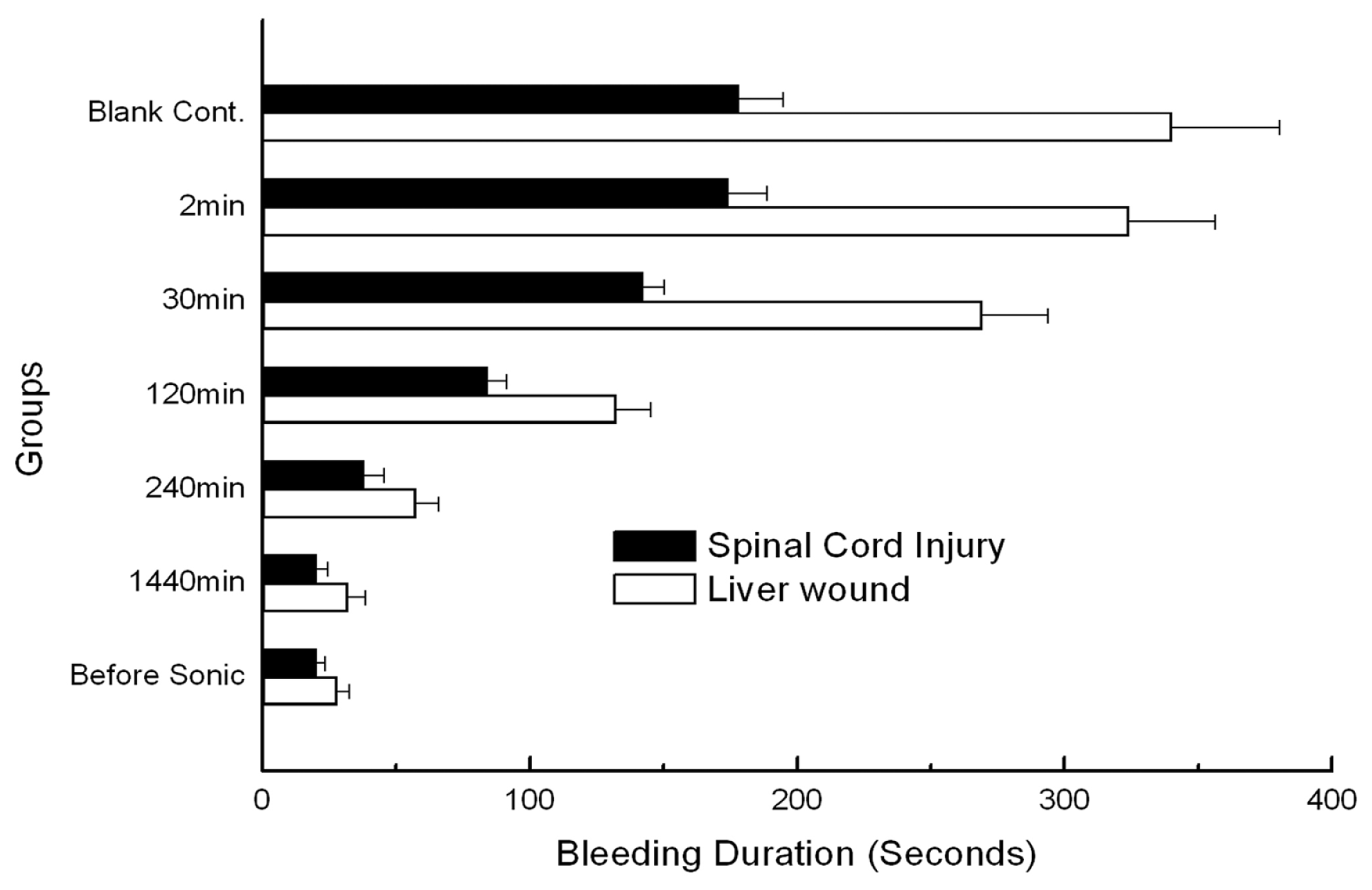
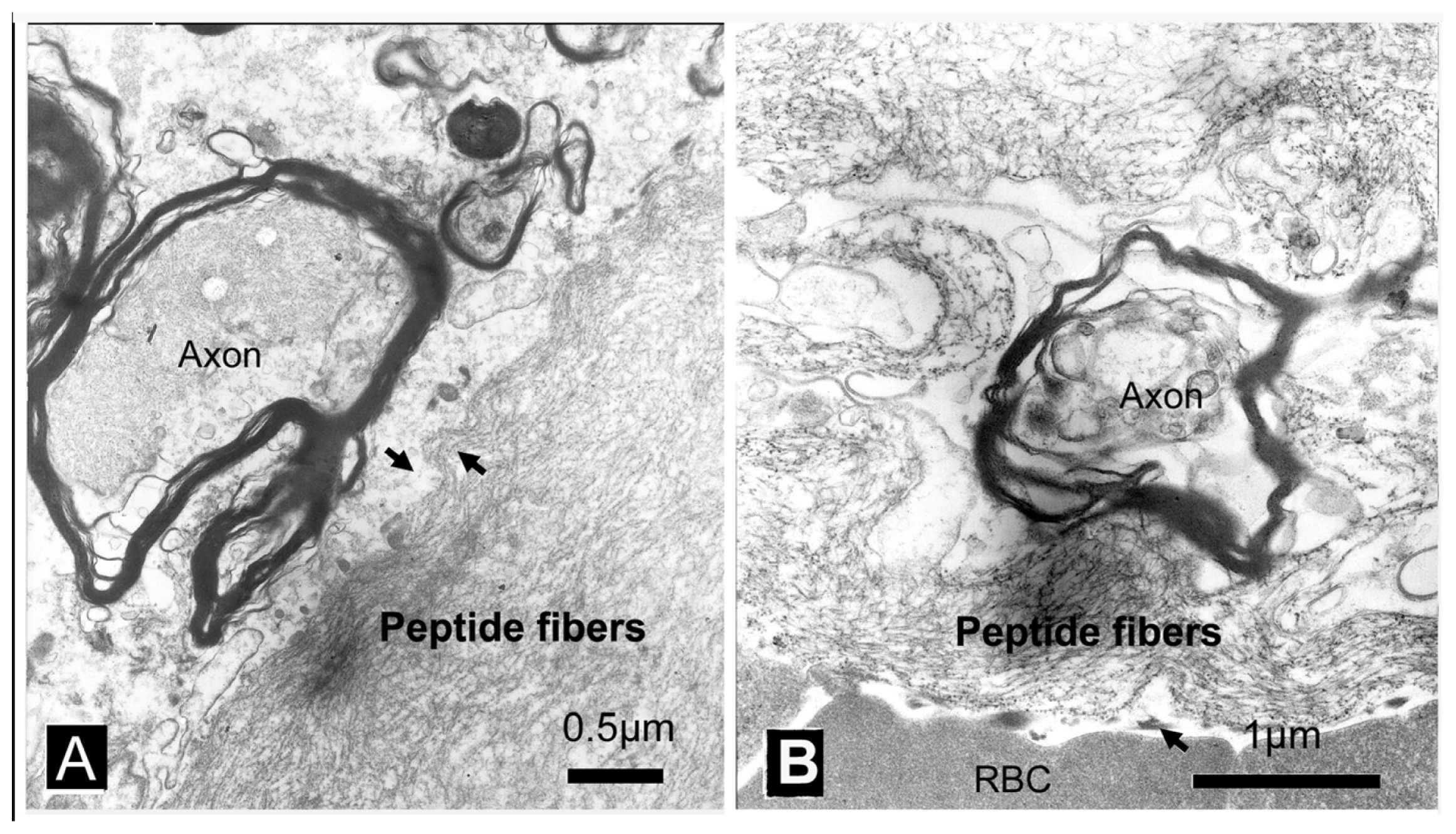
2. Results
3. Discussion
4. Experimental Procedures
4.1. Materials
4.2. Circular Dichroism (CD) Spectroscopy
4.3. Atomic Force Microscopy
4.4. Rheology Properties
4.5. Animals and Experimental Groups
4.6. Spinal Cord Transection Experiment
4.7. Liver Transverse Cut Experiments
4.8. Transmission Electron Microscopy
5. Conclusions
Acknowledgements
References
- Ellis-Behnke, R.; Liang, Y.; Tay, D.; Kau, P.; Schneider, G.; Zhang, S.; Wu, W.; So, K. Nano hemostat solution, immediate hemostasis at the nanoscale. Nanomedicine 2006, 2, 207–215. [Google Scholar]
- Alam, H.B.; Burris, D.; DaCorta, J.A.; Rhee, P. Hemorrhage control in the battlefield: Role of new hemostatic agents. Military Med 2005, 170, 63–69. [Google Scholar]
- Blocksom, J.M.; Sugawa, C.; Tokioka, S.; Williams, M. The Hemoclip: A novel approach to endoscopic therapy for esophageal perforation. Dig. Dis. Sci 2004, 49, 1136–1138. [Google Scholar]
- Kaplan, M.; Bozkurt, S.; Kut, M.S.; Kullu, S.; Demirtas, M.M. Histopathological effects of ethyl 2-cyanoacrylate tissue adhesive following surgical application: An experimental study. Eur. J. Cardio-Thorac. Surg 2004, 25, 167–172. [Google Scholar]
- Luo, Z.L.; Zhao, X.J.; Zhang, S.G. Structural dynamic of a self-assembling peptide d-EAK16 made of only D-amino acids. PLoS One 2008, 3, e2364. [Google Scholar]
- Genove, E.; Shen, C.; Zhang, S.G.; Semino, C.E. The effect of functionalized self-assembling peptide scaffolds on human aortic endothelial cell function. Biomaterials 2005, 26, 3341–3351. [Google Scholar]
- Zhang, S.G.; Holmes, T.; Lockshin, C.; Rich, A. Spontaneous assembly of a self-complementary oligopeptide to form a stable macroscopic membrane. Proc. Natl. Acad. Sci. USA 1993, 90, 3334–3338. [Google Scholar]
- Zhang, S.G.; Lockshin, C.; Cook, R.; Rich, A. Unusually stable beta-sheet formation in an ionic self-complementary oligopeptide. Biopolymers 1994, 34, 663–672. [Google Scholar]
- Ye, Z.Y.; Zhang, H.Y.; Luo, H.L.; Wang, S.K.; Zhou, Q.H.; Du, X.P.; Tang, C.K.; Chen, L.Y.; Liu, J.P.; Shi, Y.K.; et al. Temperature and pH effects on biophysical and morphological properties of self-assembling peptide RADA16-1. J. Pept. Sci 2008, 14, 152–162. [Google Scholar]
- Asakura, T.; Sugino, R.; Okumura, T.; Nakazawa, Y. The role of irregular unit, GAAS, on the secondary structure of Bombyx mori silk fibroin studied with C-13 CP/MAS NMR and wide-angle X-ray scattering. Protein Sci 2002, 11, 1873–1877. [Google Scholar]
- Wang, Y.D.; Ameer, G.A.; Sheppard, B.J.; Langer, R. A tough biodegradable elastomer. Nat. Biotechnol 2002, 20, 602–606. [Google Scholar]
- Ruan, L.P.; Zhang, H.Y.; Luo, H.L.; Liu, J.P.; Tang, F.S.; Shi, Y.K.; Zhao, X.J. Designed amphiphilic peptide forms stable nanoweb, slowly releases encapsulated hydrophobic drug, and accelerates animal hemostasis. Proc. Natl. Acad. Sci. USA 2009, 106, 5105–5110. [Google Scholar]
- Zhang, S.G. Building from bottom up: Fabrication of materials using peptide motifs. Am. Chem. Soc 2005, 229, U715–U716. [Google Scholar]
- Zhang, S.G.; Holmes, T.C.; Dipersio, C.M.; Hynes, R.O.; Su, X.; Rich, A. Self-complementary oligopeptide matrices support mammalian-cell attachment. Biomaterials 1995, 16, 1385–1393. [Google Scholar]
- Cylwik, D.; Mogielnicki, A.; Kramkowski, K.; Stokowski, J.; Buczko, W. Antithrombotic effect of l-arginine in hypertensive rats. J. Physiol. Pharmacol 2004, 55, 563–574. [Google Scholar]
- Petersen, B.; Barkun, A.; Carpenter, S.; Chotiprasidhi, P.; Chuttani, R.; Silverman, W.; Hussain, N.; Liu, J.; Taitelbaum, G.; Ginsberg, G.G. Tissue adhesives and fibrin glues. Gastrointest. Endosc 2004, 60, 327–333. [Google Scholar]
- Wang, Y.Y.; Tang, Z.Y.; Dong, M.; Liu, X.Y.; Peng, S.Q. Inhibition of platelet aggregation by polyaspartoyl l-arginine and its mechanism. Acta Pharmacol. Sinica 2004, 25, 469–473. [Google Scholar]
- Huang, W.L.; Lu, L.; Shao, X.M.; Tang, C.K.; Zhao, X.J. Anti-melanoma activity of hybrid peptide P18 and its mechanism of action. Biotechnol. Lett 2010, 32, 463–469. [Google Scholar]
- Tang, C.K.; Shao, X.M.; Sun, B.B.; Huang, W.L.; Qiu, F.; Chen, Y.Z.; Shi, Y.K.; Zhang, E.Y.; Wang, C.; Zhao, X.J. Anticancer mechanism of peptide P18 in human leukemia K562 cells. Org. Biomol. Chem 2010, 8, 984–987. [Google Scholar]
- Koutsopoulos, S.; Zhang, S. Two-layered injectable self-assembling peptide scaffold hydrogels for long-term sustained release of human antibodies. J. Control. Release 2012, 160, 451–458. [Google Scholar]
- Yokoi, H.; Kinoshita, T.; Zhang, S.G. Dynamic reassembly of peptide RADA16 nanofiber scaffold. Proc. Natl. Acad. Sci. USA 2005, 102, 8414–8419. [Google Scholar]
- Ellis-Behnke, R.G.; Liang, Y.X.; You, S.W.; Tay, D.K.C.; Zhang, S.G.; So, K.F.; Schneider, G.E. Nano neuro knitting: Peptide nanofiber scaffold for brain repair and axon regeneration with functional return of vision. Proc. Natl. Acad. Sci. USA 2006, 103, 5054–5059. [Google Scholar]
- Sharma, H.S.; Ali, S.F.; Tian, Z.R.; Hussain, S.M.; Schlager, J.J.; Sjoquist, P.O.; Sharma, A.; Muresanu, D.F. Chronic treatment with nanoparticles exacerbate hyperthermia induced blood-brain barrier breakdown, cognitive dysfunction and brain pathology in the rat. Neuroprotective effects of nanowired-antioxidant compound H-290/51. J. Nanosci. Nanotechnol 2009, 9, 5073–5090. [Google Scholar]
- Mi, K.; Wang, G.X.; Liu, Z.J.; Feng, Z.H.; Huang, B.; Zhao, X.J. Influence of a self-assembling peptide, RADA16, compared with collagen I and matrigel on the malignant phenotype of human breast-cancer cells in 3D cultures and in vivo. Macromol. Biosci 2009, 9, 437–443. [Google Scholar]
- Liu, J.P.; Song, H.; Zhang, L.L.; Xu, H.Y.; Zhao, X.J. Self-assembly-peptide hydrogels as tissue-engineering scaffolds for three-dimensional culture of chondrocytes in vitro. Macromol. Biosci 2010, 10, 1164–1170. [Google Scholar]
- Liu, X.; Wang, X.M.; Horii, A.; Wang, X.J.; Qiao, L.; Zhang, S.G.; Cui, F.Z. In vivo studies on angiogenic activity of two designer self-assembling peptide scaffold hydrogels in the chicken embryo chorioallantoic membrane. Nanoscale 2012, 4, 2720–2727. [Google Scholar]
- Meng, H.; Chen, L.Y.; Ye, Z.Y.; Wang, S.T.; Zhao, X.J. The effect of a self-assembling peptide nanofiber scaffold (peptide) when used as a wound dressing for the treatment of deep second degree burns in rats. J. Biomed. Mater. Res. B 2009, 89B, 379–391. [Google Scholar]
- Song, H.; Zhang, L.L.; Zhao, X.J. Hemostatic efficacy of biological self-assembling peptide nanofibers in a rat kidney model. Macromol. Biosci 2010, 10, 33–39. [Google Scholar]
- Qiu, F.; Chen, Y.Z.; Cheng, J.Q.; Wang, C.; Xu, H.Y.; Zhao, X.J. A simple method for cell sheet fabrication using mica surfaces grafted with peptide detergent A(6)K. Macromol. Biosci 2010, 10, 881–886. [Google Scholar]
- Wu, M.; Ye, Z.Y.; Liu, Y.F.; Liu, B.; Zhao, X.J. Release of hydrophobic anticancer drug from a newly designed self-assembling peptide. Mol. Biosyst 2011, 7, 2040–2047. [Google Scholar]
- Koutsopoulos, S.; Kaiser, L.; Eriksson, H.M.; Zhang, S.G. Designer peptide surfactants stabilize diverse functional membrane proteins. Chem. Soc. Rev 2012, 41, 1721–1728. [Google Scholar]
- Gelain, F.; Bottai, D.; Vescovi, A.; Zhang, S.G. Designer self-assembling peptide nanofiber scaffolds for adult mouse neural stem cell 3-dimensional cultures. PLoS One 2006, 1, e119. [Google Scholar]
- Sharma, H.S.; Ali, S.; Tian, Z.R.; Patnaik, R.; Patnaik, S.; Lek, P.; Sharma, A.; Lundstedt, T. Nano-drug delivery and neuroprotection in spinal cord injury. J. Nanosci. Nanotechnol 2009, 9, 5014–5037. [Google Scholar]
- Liu, J.P.; Zhang, L.L.; Yang, Z.H.; Zhao, X.J. Controlled release of paclitaxel from a self-assembling peptide hydrogel formed in situ and antitumor study in vitro. Int. J. Nanomed 2011, 6, 2143–2153. [Google Scholar]
- Lakshmanan, A.; Zhang, S.G.; Hauser, C.A.E. Short self-assembling peptides as building blocks for modern nanodevices. Trends Biotechnol 2012, 30, 155–165. [Google Scholar]



© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Wang, T.; Zhong, X.; Wang, S.; Lv, F.; Zhao, X. Molecular Mechanisms of RADA16-1 Peptide on Fast Stop Bleeding in Rat Models. Int. J. Mol. Sci. 2012, 13, 15279-15290. https://doi.org/10.3390/ijms131115279
Wang T, Zhong X, Wang S, Lv F, Zhao X. Molecular Mechanisms of RADA16-1 Peptide on Fast Stop Bleeding in Rat Models. International Journal of Molecular Sciences. 2012; 13(11):15279-15290. https://doi.org/10.3390/ijms131115279
Chicago/Turabian StyleWang, Ting, Xiaozhong Zhong, Songtao Wang, Fei Lv, and Xiaojun Zhao. 2012. "Molecular Mechanisms of RADA16-1 Peptide on Fast Stop Bleeding in Rat Models" International Journal of Molecular Sciences 13, no. 11: 15279-15290. https://doi.org/10.3390/ijms131115279
APA StyleWang, T., Zhong, X., Wang, S., Lv, F., & Zhao, X. (2012). Molecular Mechanisms of RADA16-1 Peptide on Fast Stop Bleeding in Rat Models. International Journal of Molecular Sciences, 13(11), 15279-15290. https://doi.org/10.3390/ijms131115279



