MicroRNAs as Active Players in the Pathogenesis of Multiple Sclerosis
Abstract
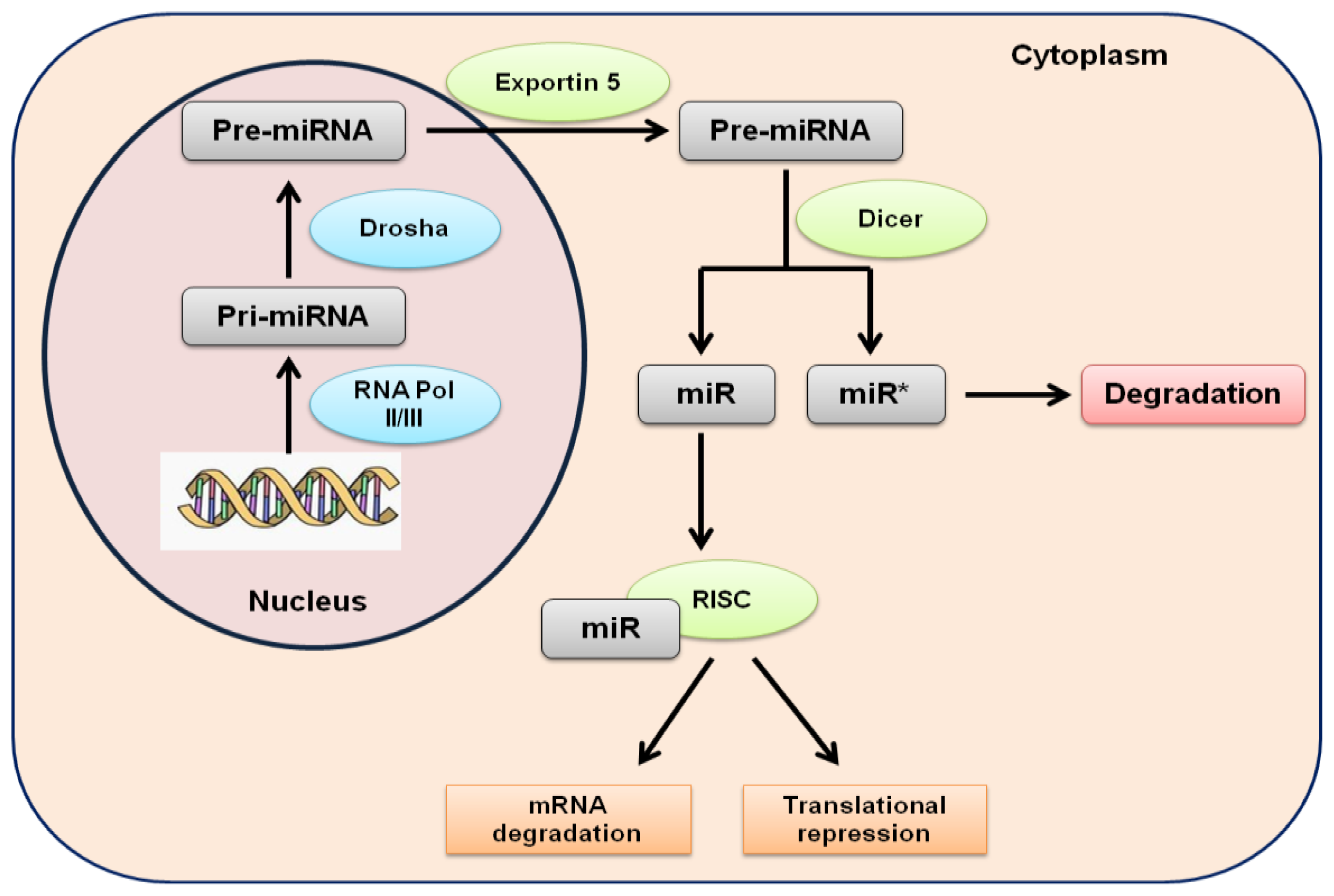
:1. Introduction
2. miRNAs and the Immune System
3. Blood and Brain Lesions miRNA Profile
4. Extracellular miRNA Profile
5. miRNAs and Genetics
6. MiRNA Therapeutic Potential
7. Treatment Effects on miRNA Profile
8. Conclusions
References
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar]
- Tufekchi, K.U.; Oner, M.G.; Genc, S.; Genc, K. MicroRNAs and multiple sclerosis. Autoimmune Diseases 2011. [Google Scholar] [CrossRef]
- Ha, T-Y. The role of microRNAs in regulatory T cells and the immune response. Immune Network 2011, 11, 11–41. [Google Scholar]
- Lages, E.; Ipas, H.; Guttin, A.; Houssan, N.; Berger, F.; Issartel, J.P. MicroRNAs: Molecular features and role in cancer. Front. Biosci 2012, 17, 2508–2540. [Google Scholar]
- O’Connell, R.M.; Rao, D.S.; Chaudhuri, A.A.; Baltimore, D. Physiological and pathological roles for microRNAs in the immune system. Nat. Rev. Immunol 2010, 10, 111–122. [Google Scholar]
- Compston, A.; Coles, A. Multiple sclerosis. Lancet 2008, 372, 1502–1517. [Google Scholar]
- Ireland, S.; Monson, N. Potential impact of B cells and T cell function in multiple sclerosis. Mult. Scler. Int 2011, 2011. [Google Scholar] [CrossRef]
- Miller, A.E. Multiple sclerosis: Where will be in 2020? Mt. Sinai J. Med 2011, 78, 268–279. [Google Scholar]
- Stys, P.K.; Zamponi, G.W.; van Minnen, J.; Geurts, J.J.G. Will the real multiple sclerosis stand up? Nat. Rev. Immunol 2012, 13, 507–514. [Google Scholar]
- Liston, A.; Linterman, M.; Lu, L-F. MicroRNA in the adaptive immune system, in sickness and in health. J. Clin. Immunol 2010, 30, 339–346. [Google Scholar]
- Tesmer, L.A.; Lundy, S.K.; Sarkar, S.; Fox, D.A. Th17 cells in human disease. Immunol. Rev 2008, 223, 87–113. [Google Scholar]
- Du, C.; Liu, C.; Kang, J.; Zhao, G.; Ye, Z.; Huang, S.; Li, Z.; Wu, Z.; Pei, G. MicroRNA miR-326 regulates Th-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat. Immunol 2009, 10, 1252–1259. [Google Scholar]
- Xiao, C.; Srinivasan, L.; Calado, D.P.; Patterson, H.C.; Zhang, B.; Wang, J.; Henderson, J.M. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat. Immunol 2008, 9, 405–414. [Google Scholar]
- Juntilla, M.M.; Koretzky, G.A. Critical roles of the PI3K/Akt signaling pathway in T cell development. Immunol. Lett 2008, 116, 104–110. [Google Scholar]
- Ovcharenco, D.; Kelnar, K.; Johnson, C.; Leng, N.; Brown, D. Genome-scale microRNA and small interfering RNA screens identify small RNA modulators of TRAIL-induced apoptosis pathway. Cancer Res 2007, 67, 10782–10788. [Google Scholar]
- Lindberg, R.L.P.; Hoffmann, F.; Mehling, M.; Kuhle, J.; Kappos, L. Altered expression of miR-17-5p in CD4+ lymphocytes of relapsing-remitting multiple sclerosis patients. Eur. J. Immunol 2010, 40, 888–898. [Google Scholar]
- Tang, Q.; Bluestone, J.A. Regulatory T cell physiology and application to treat autoimmunity. Immunol. Rev 2006, 212, 217–237. [Google Scholar]
- Petrocca, F.; Vecchione, A.; Croce, C.M. Emerging role of miR-106b-25/miR-17-92 clusters in the control of transforming growth factor beta signaling. Cancer Res 2008, 68, 8191–8194. [Google Scholar]
- De Santis, G.; Ferracin, M.; Biondani, A.; Caniatti, L.; Tola, M.A.; Castellezzi, M.; Zagatti, B.; Battistini, L.; Borsellino, G.; Fainardi, E.; et al. Altered miRNA expression in T regulatory cells in course of multiple sclerosis. J. Neuroimmnuol 2010, 226, 165–171. [Google Scholar]
- Guerau-de-Arellano, M.; Smith, K.M.; Godlewski, J.; Liu, Y.; Winger, R.; Lawler, S.E.; Whitacre, C.C.; Racke, M.K.; Lovett-Racke, A.E. Micro-RNA dysregulation in multiple sclerosis favours pro-inflammatory T-cell-mediated autoimmunity. Brain 2011, 134, 3578–3589. [Google Scholar]
- Otaegui, D.; Baranzini, S.E.; Armananzas, R.; Calvo, B.; Munoz-Culla, M.; Khankhanian, P.; Inza, I.; Lozano, J.A.; Castillo-Trivino, T.; Asensio, A.; et al. Differential micro RNA expression in PBMC from multiple sclerosis patients. PLoS One 2009, 4. [Google Scholar] [CrossRef] [Green Version]
- Fenoglio, C.; Cantoni, C.; de Riz, M.; Ridolfi, E.; Cortini, F.; Sepente, M.; Villa, C.; Comi, C.; Monaco, F.; Mellesi, L.; et al. Expression and genetic analysis of miRNAs involved in CD4+ cell activation in patients with multiple sclerosis. Neurosci. Lett 2011, 504, 9–12. [Google Scholar]
- Martinelli-Boneschi, F.; Fenoglio, C.; Brambilla, P.; Sorosina, M.; Giacalone, G.; Esposito, F.; Serpente, M.; Cantoni, C.; Ridolfi, E.; Rodegher, M.; et al. MicroRNA and mRNA expression profile screening in multiple sclerosis patients to unravel novel pathogenic steps and identify potential biomarkers. Neurosci. Lett 2012, 508, 4–8. [Google Scholar]
- Keller, A.; Leidinger, P.; Lange, J.; Borries, A.; Schroers, H.; Scheffler, M.; Lenhof, H.P.; Ruprecht, K.; Meese, E. Multiple sclerosis: MicroRNA expression profiles accurately differentiate patients with relapsing-remitting disease from healthy controls. PLoS One 2009, 4. [Google Scholar] [CrossRef]
- Cox, M.B.; Cairns, M.J.; Gandhi, K.S.; Carroll, A.P.; Moscovis, S.; Stewart, G.J.; Broadley, S.; Scott, R.J.; Booth, D.R.; Lechner-Scott, J. ANZgene Multiple Sclerosis Genetics Consortium. MicroRNAs miR-17 and miR-20a inhibit T cell activation genes and are under-expressed in MS whole blood. PLoS One 2010, 5. [Google Scholar] [CrossRef]
- Junker, A.; Krumbholz, M.; Eisele, S.; Mohan, H.; Augstein, F.; Bittner, R.; Lassmann, H.; Wekerle, H.; Hohlfeld, R.; Meinl, E. MicroRNA profiling of multiple sclerosis lesions identifies modulators of the regulatory protein CD47. Brain 2009, 132, 3342–3352. [Google Scholar]
- Hanke, M.; Hoefig, K.; Merz, H.; Feller, A.C.; Kausch, I. A robust methodology to study urine microRNA as tumor marker: MicroRNA-126 and microRNA-182 are related tourinary bladder cancer. Urol. Oncol 2010, 28, 655–661. [Google Scholar]
- Park, N.J.; Zhou, H.; Elashoff, D.; Henson, B.S.; Kastratovic, D.A. Salivary microRNA: Discovery, characterization, and clinical utility for oral cancer detection. Clin. Cancer Res 2009, 15, 5473–5477. [Google Scholar]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar]
- Redell, J.B.; Moore, A.N.; Ward, N.H., III; Hergenroeder, G.W.; Dash, P.K. Human traumatic brain injury alters plasma microRNA levels. J. Neurotrauma 2009, 27, 2147–2156. [Google Scholar]
- Boeri, M.; Verri, C.; Conte, D.; Roz, L.; Modena, P. MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc. Natl. Acad. Sci. USA 2011, 108, 3713–3718. [Google Scholar]
- Siegel, S.R.; Mackenzie, J.; Chaplin, G.; Jablonski, N.G.; Griffiths, L. Circulating microRNAs involved in multiple sclerosis. Mol. Biol. Rep 2012, 39, 6219–6225. [Google Scholar]
- Fenoglio, C.; Ridolfi, E.; Serpente, M.; Cantoni, C.; de Riz, M.; Pietroboni, A.; Villa, C.; Cortini, F.; Piccio, L.; Bresolin, N.; et al. Cell-free microRNA and multiple sclerosis: Possible promising biomarkers? Mult. Scler 2011, 17, S53–S276. [Google Scholar]
- Clemente, D.; Ortega, M.C.; Arenzana, F.J.; de Castro, F. FGF-2 and Anosmin-1 are selectively expressed in different types of multiple sclerosis lesions. J. Neurosci 2011, 31, 14899–14909. [Google Scholar]
- Sarchielli, P.; di Filippo, M.; Ercolani, M.V.; Chiasserini, D.; Mattioni, A. Fibroblast growth factor-2 levels are elevated in the cerebrospinal fluid of multiple sclerosis patients. Neurosci. Lett 2008, 435, 223–228. [Google Scholar]
- Aulchenko, Y.S.; Hoppenbrouwers, I.A.; Ramagopalan, S.V.; Broer, L.; Jafari, N. Genetic variation in the KIF1B locus influences susceptibility to multiple sclerosis. Nat Genet 2008, 40, 1402–1403. [Google Scholar]
- Lyons, D.A.; Naylor, S.G.; Scholze, A.; Talbot, W.S. Kif1b is essential for mRNA localization in oligodendrocytes and development of myelinated axons. Nat. Genet 2009, 41, 854–858. [Google Scholar]
- Martinelli-Boneschi, F.; Esposito, F.; Scalabrini, D.; Fenoglio, C.; Rodegher, M.E. Lack of replication of KIF1B gene in an Italian primary progressive multiple sclerosis cohort. Eur. J. Neurol 2010, 17, 740–745. [Google Scholar]
- Goris, A.; Boonen, S.; D’hooghe, M.B.; Dubois, B. Replication of KIF21B as a susceptibility locus for multiple sclerosis. J. Med. Genet 2010, 47, 775–776. [Google Scholar]
- Koutsis, G.; Karadima, G.; Floroskufi, P.; Sfagos, C.; Vassilopoulos, D.; Panas, M. The rs10492972 KIF1B polymorphism and disease progression in Greek patients with multiple sclerosis. J. Neurol 2011, 258, 1726–1728. [Google Scholar]
- Sombekke, M.H.; Jafari, N.; Bendfeldt, K.; Mueller-Lenke, N.; Radue, E.W. No influence of KIF1B on neurodegenerative markers in multiple sclerosis. Neurology 2011, 76, 1843–1845. [Google Scholar]
- Kudryavtseva, E.A.; Rozhdestvenskii, A.S.; Kakulya, A.V.; Khanokh, E.V.; Delov, R.A. Polymorphic locus rs10492972 of the KIF1B gene association with multiple sclerosis in Russia: Case control study. Mol. Genet. Metab 2011, 104, 390–394. [Google Scholar]
- Nowakowska, B.A.; Obersztyn, E.; Szymańska, K.; Bekiesińska-Figatowska, M.; Xia, Z. Severe mental retardation, seizures, and hypotonia due to deletions of MEF2C. Am. J. Med. Genet. Part B 2010, 153, 1042–1051. [Google Scholar]
- Li, T.; Morgan, M.J.; Choksi, S.; Zhang, Y.; Kim, Y.S.; Liu, Z.G. MicroRNAs modulate the noncanonical transcription factor NF-κB pathway by regulating expression of the kinase IKKα during macrophage differentiation. Nat. Immunol 2010, 11, 799–805. [Google Scholar]
- Fulci, V.; Scappucci, G.; Sebastiani, G.D.; Giannitti, C.; Franceschini, D. miR-223 is overexpressed in T-lymphocytes of patients affected by rheumatoid arthritis. Hum. Immunol 2010, 71, 206–211. [Google Scholar]
- Hoppenbrouwers, I.A.; Hintzen, R. Genetics of multiple sclerosis. Biochim. Biophys. Acta 2011, 1812, 194–201. [Google Scholar]
- Gourraud, P.A.; Harbo, H.F.; Hauser, S.L.; Baranzini, S.E. The genetics of multiple sclerosis: An up-to-date review. Immunol. Rev 2012, 248, 87–103. [Google Scholar]
- Salzman, D.W.; Weidhaas, J.B. SNPing cancer in the bud: MicroRNA and microRNA-target site polymorphisms as diagnostic and prognostic biomarkers in cancer. Pharmacol. Ther 2012, in press. [Google Scholar]
- Paraboschi, E.M.; Soldà, G.; Gemmati, D.; Orioli, E.; Zeri, G.; Benedetti, M.D.; Salviati, A.; Barizzone, N.; Leone, M.; Duga, S.; et al. Genetic association and altered gene expression of miR-155 in multiple sclerosis patients. Int. J. Mol. Sci 2011, 12, 8695–8712. [Google Scholar]
- Sonkoly, E.; Wei, T.; Janson, P.C.; Saaf, A.; Lundeberg, L.; Tengvall-Linder, M.; Norstedt, G.; Alenius, H.; Homey, B.; Scheynius, A.; et al. MicroRNAs: Novel regulators involved in the pathogenesis of psoriasias? PLoS One 2007, 2. [Google Scholar] [CrossRef]
- Cobb, B.S.; Hertweck, A.; Smith, J.; O’Connor, E.; Graf, D.; Cook, T.; Smale, S.T.; Sakaguchi, S.; Livesey, F.J.; Fisher, A.G.; et al. A role for Dicer in immune regulation. J. Exp. Med 2006, 203, 2519–2527. [Google Scholar]
- Murugaiyan, G.; Beynon, V.; Mittal, A.; Joller, N.; Weiner, H.L. Silencing microRNA-155 ameliorates experimental autoimmune encephalomyelitis. J. Immunol 2011, 187, 2213–2221. [Google Scholar]
- Czech, M. MicroRNAs as therapeutic targets. N. Engl. J. Med 2006, 354, 1194–1195. [Google Scholar]
- Krützfeldt, J.; Rajewsky, N.; Braich, R.; Rajeev, K.G.; Tuschl, T.; Manoharan, M.; Stoffel, M. Silencing of microRNAs in vivo with “antagomirs”. Nature 2005, 438, 685–689. [Google Scholar]
- Im, H.I.; Kenny, P.J. MicroRNAs in neuronal function and dysfunction. Trends Neurosci 2012, 35, 325–334. [Google Scholar]
- Li, Y.; He, C.; Jin, P. Emergence of chemical biology approaches to the RNAi/miRNA pathway. Chem. Biol 2010, 17, 584–589. [Google Scholar]
- Watashi, K. Identification of small molecules that suppress microRNA function and reverse tumorigenesis. J. Biol. Chem 2010, 285, 24707–24716. [Google Scholar]
- Steinman, L. A rush to judgment on Th17. J. Exp. Med 2008, 205, 1517–1522. [Google Scholar]
- Tzartos, J.S.; Friese, M.A.; Craner, M.J.; Palace, J.; Newcombe, J.; Esiri, M.M.; Fugger, L. Interleukin-17 production in central nervous system-infiltrating T cells and glial cells is associated with active disease in multiple sclerosis. Am. J. Pathol 2008, 172, 146–155. [Google Scholar]
- Waschbisch, A.; Atiya, M.; Linker, R.A.; Potapov, S.; Schwab, S.; Derfuss, T. Glatiramer acetate treatment normalizes deregulated microRNA expression in relapsing remitting multiple sclerosis. PLoS One 2011, 6. [Google Scholar] [CrossRef]
- Sievers, C.; Meira, M.; Hoffmann, F.; Fontoura, P.; Kappos, L.; Lindberg, R.L. Altered microRNA expression in B lymphocytes in multiple sclerosis: Towards a better understanding of treatment effects. Clin. Immunol 2012, 144, 70–79. [Google Scholar]
© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Fenoglio, C.; Ridolfi, E.; Galimberti, D.; Scarpini, E. MicroRNAs as Active Players in the Pathogenesis of Multiple Sclerosis. Int. J. Mol. Sci. 2012, 13, 13227-13239. https://doi.org/10.3390/ijms131013227
Fenoglio C, Ridolfi E, Galimberti D, Scarpini E. MicroRNAs as Active Players in the Pathogenesis of Multiple Sclerosis. International Journal of Molecular Sciences. 2012; 13(10):13227-13239. https://doi.org/10.3390/ijms131013227
Chicago/Turabian StyleFenoglio, Chiara, Elisa Ridolfi, Daniela Galimberti, and Elio Scarpini. 2012. "MicroRNAs as Active Players in the Pathogenesis of Multiple Sclerosis" International Journal of Molecular Sciences 13, no. 10: 13227-13239. https://doi.org/10.3390/ijms131013227



