MiRNAs and LincRNAs: Could They Be Considered as Biomarkers in Colorectal Cancer?
Abstract
:1. Introduction
2. miRNAs and lincRNA Mechanism of Action
3. miRNAs, lincRNA and Cancer
4. miRNA Expression in CRC
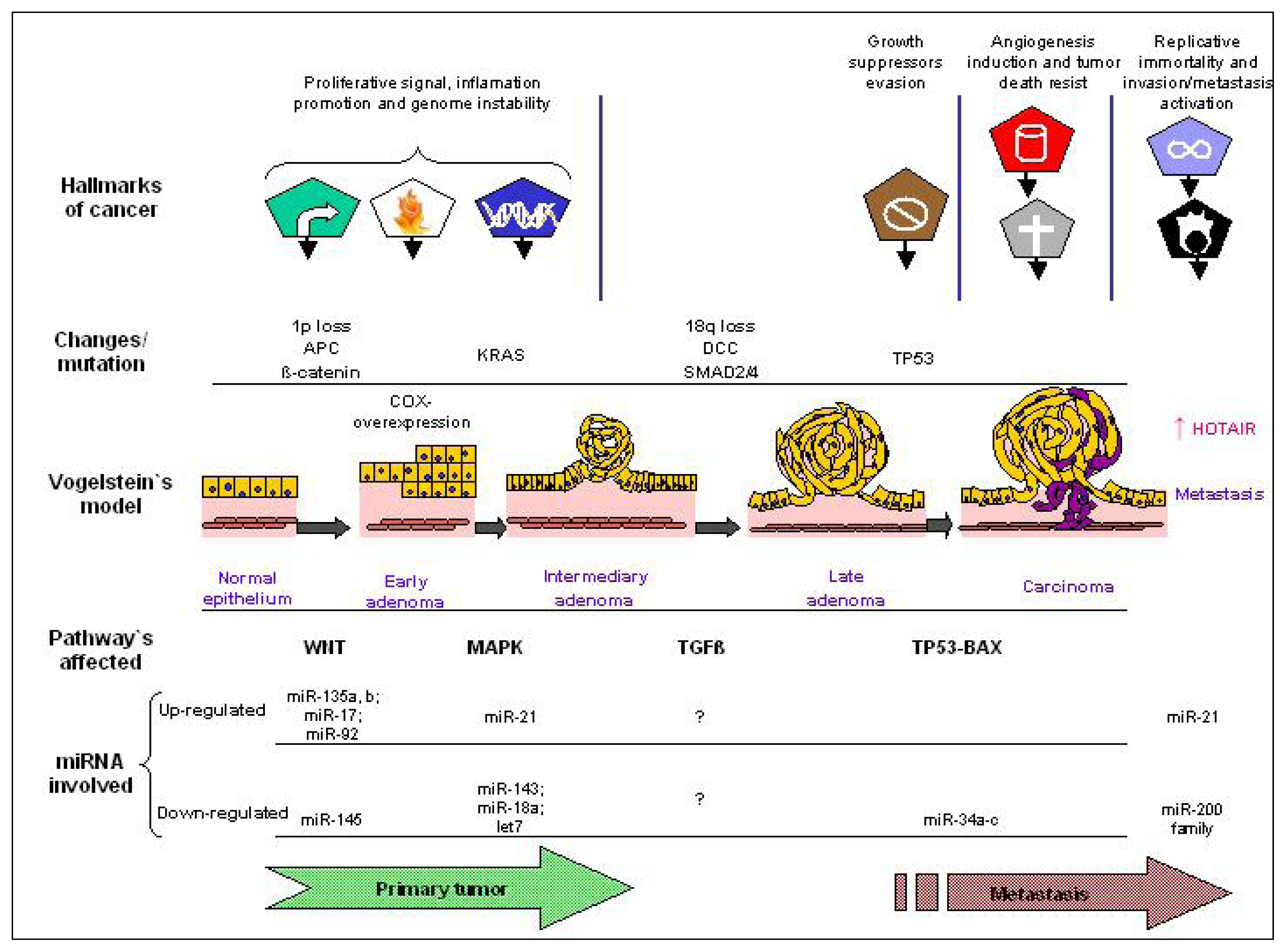
5. ncRNA as Regulator of Tumor Hallmarkers and Vogelstein’s Model of CRC Pathogenesis
6. miRNA and Chemoresistance
7. SNPs and miRNA
8. miRNAs as Predictive and Prognostic Biomarkers in CRC
9. miRNAs as “One Plus” Potential Cancer Therapeutic Targets
10. Summary
References
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar]
- Wightman, B.; Ha, I.; Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993, 75, 855–862. [Google Scholar]
- Filipowicz, W.; Bhattacharyya, S.N.; Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet 2008, 9, 102–114. [Google Scholar]
- Perez, D.S.; Hoage, T.R.; Pritchett, J.R.; Ducharme-Smith, A.L.; Halling, M.L.; Ganapathiraju, S.C.; Streng, P.S.; Smith, D.I. Long, abundantly expressed noncoding transcripts are altered in cancer. Hum. Mol. Genet 2008, 17, 642–655. [Google Scholar]
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P.; et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009, 458, 223–227. [Google Scholar]
- Lagos-Quintana, M.; Rauhut, R.; Lendeckel, W.; Tuschl, T. Identification of novel genes coding for small expressed RNAs. Science 2001, 294, 853–858. [Google Scholar]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar]
- Bartel, D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar]
- John, B.; Enright, A.J.; Aravin, A.; Tuschl, T.; Sander, C.; Marks, D.S. Human MicroRNA targets. PLoS Biol 2004, 2, e363. [Google Scholar]
- Cheng, J.; Kapranov, P.; Drenkow, J.; Dike, S.; Brubaker, S.; Patel, S.; Long, J.; Stern, D.; Tammana, H.; Helt, G.; et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science 2005, 308, 1149–1154. [Google Scholar]
- Wu, Q.; Kim, Y.C.; Lu, J.; Xuan, Z.; Chen, J.; Zheng, Y.; Zhou, T.; Zhang, M.Q.; Wu, C.I.; Wang, S.M. Poly A- transcripts expressed in HeLa cells. PLoS One 2008, 3, e2803. [Google Scholar]
- Okazaki, Y.; Furuno, M.; Kasukawa, T.; Adachi, J.; Bono, H.; Kondo, S.; Nikaido, I.; Osato, N.; Saito, R.; Suzuki, H.; et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 2002, 420, 563–573. [Google Scholar]
- Khalil, A.M.; Guttman, M.; Huarte, M.; Garber, M.; Raj, A.; Rivea Morales, D.; Thomas, K.; Presser, A.; Bernstein, B.E.; van Oudenaarden, A.; et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. USA 2009, 106, 11667–11672. [Google Scholar]
- Brannan, C.I.; Dees, E.C.; Ingram, R.S.; Tilghman, S.M. The product of the H19 gene may function as RNA. Mol. Cell. Biol 1990, 10, 28–36. [Google Scholar]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar]
- Willingham, A.T.; Orth, A.P.; Batalov, S.; Peters, E.C.; Wen, B.G.; Aza-Blanc, P.; Hogenesch, J.B.; Schultz, P.G. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science 2005, 309, 1570–1573. [Google Scholar]
- Mercer, T.R.; Dinger, M.E.; Sunkin, S.M.; Mehler, M.F.; Mattick, J.S. Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl. Acad. Sci. USA 2008, 105, 716–721. [Google Scholar]
- Johnson, S.M.; Grosshans, H.; Shingara, J.; Byrom, M.; Jarvis, R.; Cheng, A.; Labourier, E.; Reinert, K.L.; Brown, D.; Slack, F.J. RAS is regulated by the let-7 microRNA family. Cell 2005, 120, 635–647. [Google Scholar]
- Lee, Y.S.; Dutta, A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev 2007, 21, 1025–1030. [Google Scholar]
- Sampson, V.B.; Rong, N.H.; Han, J.; Yang, Q.; Aris, V.; Soteropoulos, P.; Petrelli, N.J.; Dunn, S.P.; Krueger, L.J. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res 2007, 67, 9762–9770. [Google Scholar]
- Lan, F.F.; Wang, H.; Chen, Y.C.; Chan, C.Y.; Ng, S.S.; Li, K.; Xie, D.; He, M.L.; Lin, M.C.; Kung, H.F. Hsa-let-7g inhibits proliferation of hepatocellular carcinoma cells by downregulation of c-Myc and upregulation of p16 (INK4A). Int. J. Cancer 2011, 128, 319–331. [Google Scholar]
- Corney, D.C.; Flesken-Nikitin, A.; Godwin, A.K.; Wang, W.; Nikitin, A.Y. MicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res 2007, 67, 8433–8438. [Google Scholar]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: the next generation. Cell 2011, 144, 646–674. [Google Scholar]
- Gregory, R.I.; Chendrimada, T.P.; Cooch, N.; Shiekhattar, R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 2005, 123, 631–640. [Google Scholar]
- Hutvagner, G. Small RNA asymmetry in RNAi: function in RISC assembly and gene regulation. FEBS Lett 2005, 579, 5850–5857. [Google Scholar]
- Lim, L.P.; Lau, N.C.; Garrett-Engele, P.; Grimson, A.; Schelter, J.M.; Castle, J.; Bartel, D.P.; Linsley, P.S.; Johnson, J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 2005, 433, 769–773. [Google Scholar]
- Grimson, A.; Farh, K.K.; Johnston, W.K.; Garrett-Engele, P.; Lim, L.P.; Bartel, D.P. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell 2007, 27, 91–105. [Google Scholar]
- Maziere, P.; Enright, A.J. Prediction of microRNA targets. Drug Discov. Today 2007, 12, 452–458. [Google Scholar]
- Bartonicek, N.; Enright, A.J. SylArray: A web server for automated detection of miRNA effects from expression data. Bioinformatics 2010, 26, 2900–2911. [Google Scholar]
- Heikham, R.; Shankar, R. Flanking region sequence information to refine microRNA target predictions. J. Biosci 2010, 35, 105–118. [Google Scholar]
- Maragkakis, M.; Reczko, M.; Simossis, V.A.; Alexiou, P.; Papadopoulos, G.L.; Dalamagas, T.; Giannopoulos, G.; Goumas, G.; Koukis, E.; Kourtis, K.; et al. DIANA-microT web server: Elucidating microRNA functions through target prediction. Nucleic Acids Res 2009, 37, W273–276. [Google Scholar]
- Kertesz, M.; Iovino, N.; Unnerstall, U.; Gaul, U.; Segal, E. The role of site accessibility in microRNA target recognition. Nat. Genet 2007, 39, 1278–1284. [Google Scholar]
- Thadani, R.; Tammi, M.T. MicroTar: Predicting microRNA targets from RNA duplexes. BMC Bioinformatics 2006, 7, S20. [Google Scholar]
- Kim, S.K.; Nam, J.W.; Rhee, J.K.; Lee, W.J.; Zhang, B.T. miTarget: microRNA target gene prediction using a support vector machine. BMC Bioinformatics 2006, 7, 411. [Google Scholar]
- Miranda, K.C.; Huynh, T.; Tay, Y.; Ang, Y.S.; Tam, W.L.; Thomson, A.M.; Lim, B.; Rigoutsos, I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell 2006, 126, 1203–1217. [Google Scholar]
- Kruger, J.; Rehmsmeier, M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res 2006, 34, W451–W454. [Google Scholar]
- Krek, A.; Grün, D.; Poy, M.N.; Wolf, R.; Rosenberg, L.; Epstein, E.J.; MacMenamin, P.; da Piedade, I.; Gunsalus, K.C.; Stoffel, M.; et al. Combinatorial microRNA target predictions. Nat. Genet 2005, 37, 495–500. [Google Scholar]
- John, B.; Enright, A.J.; Aravin, A.; Tuschl, T.; Sander, C.; Marks, D.S. Human MicroRNA targets. PLoS Biol 2004, 2, e363. [Google Scholar]
- Lewis, B.P.; Shih, I.H.; Jones-Rhoades, M.W.; Bartel, D.P.; Burge, C.B. Prediction of mammalian microRNA targets. Cell 2003, 115, 787–798. [Google Scholar]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar]
- Li, L.; Xu, J.; Yang, D.; Tan, X.; Wang, H. Computational approaches for microRNA studies: A review. Mamm. Genome 2010, 21, 1–12. [Google Scholar]
- Huarte, M.; Rinn, J.L. Large non-coding RNAs: Missing links in cancer? Hum. Mol. Genet 2010, 19, 152–161. [Google Scholar]
- MiRBase. Available online: http://microrna.sanger.ac.uk/sequences/index.shtml accessed on 16 January 2012.
- MiRBase. Available online: http://www.miRBase.org/ accessed on 16 January 2012.
- Zhang, L.; Huang, J.; Yang, N.; Greshock, J.; Megraw, M.S.; Giannakakis, A.; Liang, S.; Naylor, T.L.; Barchetti, A.; Ward, M.R.; et al. MicroRNAs exhibit high frequency genomic alterations in human cancer. Proc. Natl. Acad. Sci. USA 2006, 103, 9136–9141. [Google Scholar]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–1559. [Google Scholar]
- Cimmino, A.; Calin, G.A.; Fabbri, M.; Iorio, M.V.; Ferracin, M.; Shimizu, M.; Wojcik, S.E.; Aqeilan, R.I.; Zupo, S.; Dono, M.; et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. USA 2005, 102, 13944–13949. [Google Scholar]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar]
- Volinia, S.; Calin, G.A.; Liu, C.G.; Ambs, S.; Cimmino, A.; Petrocca, F.; Visone, R.; Iorio, M.; Roldo, C.; Ferracin, M.; et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 2006, 103, 2257–2261. [Google Scholar]
- Calin, G.A.; Croce, C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer 2006, 6, 857–866. [Google Scholar]
- Cho, W.C. MicroRNAs: Potential biomarkers for cancer diagnosis, prognosis and targets for therapy. Int. J. Biochem. Cell Biol 2010, 42, 1273–1281. [Google Scholar]
- Cho, W.C. MicroRNAs in cancer—from research to therapy. Biochim. Biophys. Acta 2010, 1805, 209–217. [Google Scholar]
- Luo, X.; Burwinkel, B.; Tao, S.; Brenner, H. MicroRNA signatures: Novel biomarker for colorectal cancer? Cancer Epidemiol. Biomarkers Prev 2011, 20, 1272–1286. [Google Scholar]
- Katayama, S.; Tomaru, Y.; Kasukawa, T.; Waki, K.; Nakanishi, M.; Nakamura, M.; Nishida, H.; Yap, C.C.; Suzuki, M.; Kawai, J.; et al. Antisense transcription in the mammalian transcriptome. Science 2005, 309, 1564–1566. [Google Scholar]
- Khalil, A.M.; Guttman, M.; Huarte, M.; Garber, M.; Raj, A.; Rivea Morales, D.; Thomas, K.; Presser, A.; Bernstein, B.E.; van Oudenaarden, A.; et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. USA 2009, 106, 11667–11672. [Google Scholar]
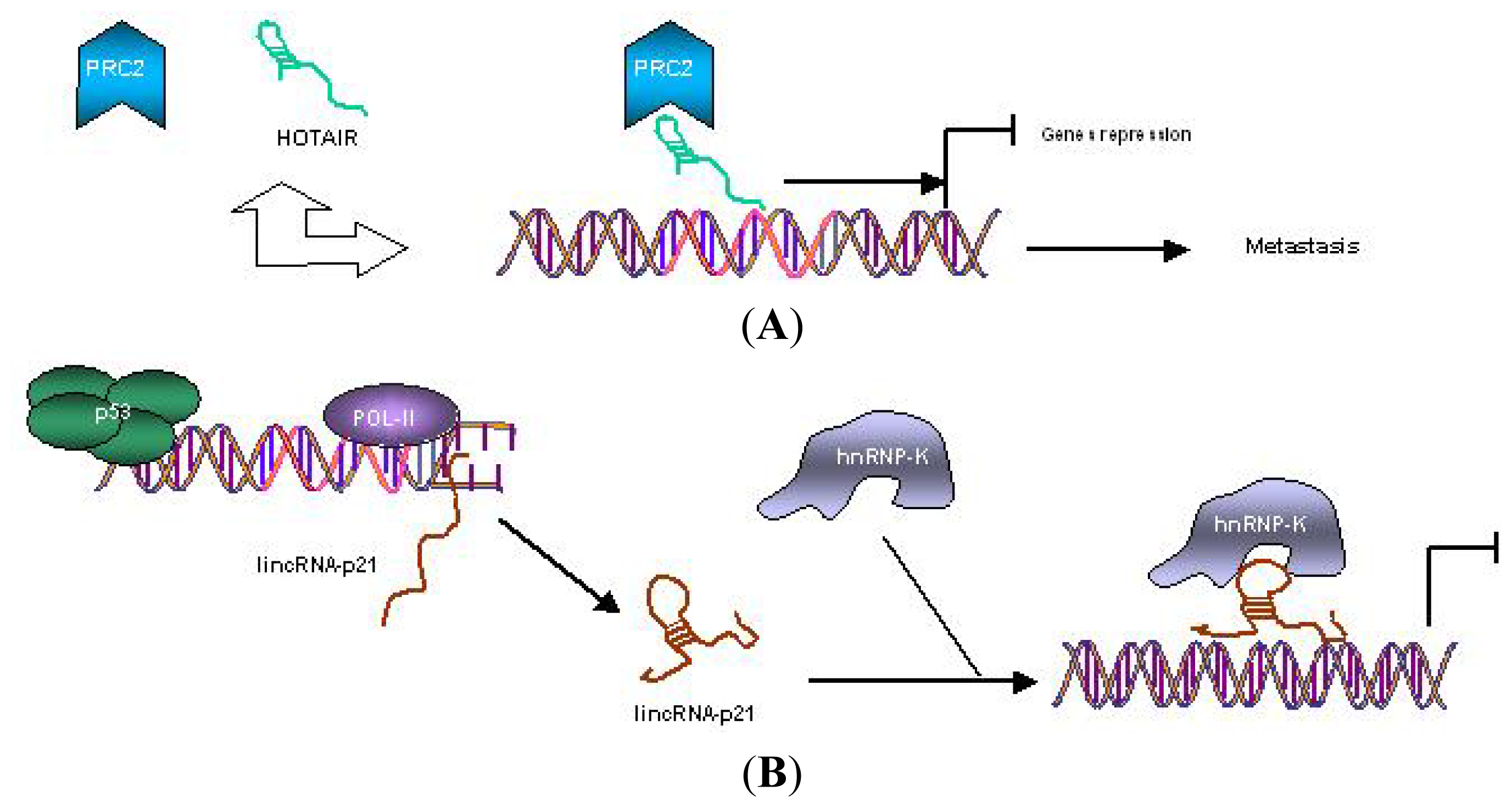
- Huarte, M.; Guttman, M.; Feldser, D.; Garber, M.; Koziol, M.J.; Kenzelmann-Broz, D.; Khalil, A.M.; Zuk, O.; Amit, I.; Rabani, M.; et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 2010, 142, 409–419. [Google Scholar]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar]
- Michael, M.Z.; O’Connor, S.M.; van Holst Pellekaan, N.G.; Young, G.P.; James, R.J. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol. Cancer Res 2003, 1, 882–891. [Google Scholar]
- Cummins, J.M.; He, Y.; Leary, R.J.; Pagliarini, R.; Diaz, L.A., Jr.; Sjoblom, T.; Barad, O.; Bentwich, Z.; Szafranska, A.E.; Labourier, E.; et al. The colorectal microRNAome. Proc. Natl. Acad. Sci. USA 2006, 103, 3687–3692. [Google Scholar]
- Bandrés, E.; Cubedo, E.; Agirre, X.; Malumbres, R.; Zárate, R.; Ramirez, N.; Abajo, A.; Navarro, A.; Moreno, I.; Monzó, M.; et al. Identification by Real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol. Cancer 2006, 5, 29. [Google Scholar]
- Pawa, N.; Arulampalam, T.; Norton, J.D. Screening for colorectal cancer: Established and emerging modalities. Nat. Rev. Gastroenterol. Hepatol 2011, 8, 711–722. [Google Scholar]
- Fearon, E.R.; Vogelstein, B. A genetic model for colorectal tumorigenesis. Cell 1990, 61, 759–767. [Google Scholar]
- Nagel, R.; le Sage, C.; Diosdado, B.; van der Waal, M.; Oude Vrielink, J.A.; Bolijn, A.; Meijer, G.A.; Agami, R. Regulation of the adenomatous polyposis coli gene by the miR-135 family in colorectal cancer. Cancer Res 2008, 68, 5795–5802. [Google Scholar]
- Kumar, M.S.; Lu, J.; Mercer, K.L.; Golub, T.R.; Jacks, T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat. Genet 2007, 39, 673–677. [Google Scholar]
- Tsang, W.P.; Kwok, T.T. The miR-18a* microRNA functions as a potential tumor suppressor by targeting on K-Ras. Carcinogenesis 2009, 30, 953–959. [Google Scholar]
- Guo, C.; Sah, J.F.; Beard, L.; Willson, J.K.; Markowitz, S.D.; Guda, K. The noncoding RNA, miR-126, suppresses the growth of neoplastic cells by targeting phosphatidylinositol 3-kinase signaling and is frequently lost in colon cancers. Genes Chromosomes Cancer 2008, 47, 939–946. [Google Scholar]
- Natalwala, A.; Spychal, R.; Tselepis, C. Epithelial-mesenchymal transition mediated tumourigenesis in the gastrointestinal tract. World J. Gastroenterol 2008, 14, 3792–3797. [Google Scholar]
- Strillacci, A.; Griffoni, C.; Sansone, P.; Paterini, P.; Piazzi, G.; Lazzarini, G.; Spisni, E.; Pantaleo, M.A.; Biasco, G.; Tomasi, V. MiR-101 downregulation is involved in cyclooxygenase-2 overexpression in human colon cancer cells. Exp. Cell Res 2009, 315, 1439–1447. [Google Scholar]
- Hudson, J.D.; Shoaibi, M.A.; Maestro, R.; Carnero, A.; Hannon, G.J.; Beach, D.H. A proinflammatory cytokine inhibits p53 tumor suppressor activity. J. Exp. Med 1999, 190, 1375–1382. [Google Scholar]
- Ohkawara, T.; Takeda, H.; Nishihira, J.; Miyashita, K.; Nihiwaki, M.; Ishiguro, Y.; Takeda, K.; Akira, S.; Iwanaga, T.; Sugiyama, T.; et al. Macrophage migration inhibitory factor contributes to the development of acute dextran sulphate sodium-induced colitis in Toll-like receptor 4 knockout mice. Clin. Exp. Immunol 2005, 141, 412–421. [Google Scholar]
- Wilson, J.M.; Coletta, P.L.; Cuthbert, R.J.; Scott, N.; MacLennan, K.; Hawcroft, G.; Leng, L.; Lubetsky, J.B.; Jin, K.K.; Lolis, E.; et al. Macrophage migration inhibitory factor promotes intestinal tumorigenesis. Gastroenterology 2005, 129, 1485–1503. [Google Scholar]
- He, X.X.; Chen, K.; Yang, J.; Li, X.Y.; Gan, H.Y.; Liu, C.Y.; Coleman, T.R.; Al-Abed, Y. Macrophage migration inhibitory factor promotes colorectal cancer. Mol. Med 2009, 15, 1–10. [Google Scholar]
- Shah, M.A.; Schwartz, G.K. Cell cycle-mediated drug resistance: an emerging concept in cancer therapy. Clin. Cancer Res 2001, 7, 2168–2181. [Google Scholar]
- Mirjolet, J.F.; Barberi-Heyob, M.; Merlin, J.L.; Marchal, S.; Etienne, M.C.; Milano, G.; Bey, P. Thymidylate synthase expression and activity: Relation to S-phase parameters and 5-fluorouracil sensitivity. Br. J. Cancer 1998, 78, 62–68. [Google Scholar]
- McGinn, C.J.; Pestalozzi, B.C.; Drake, J.C.; Glennon, M.C.; Kunugi, K.; Otterson, G.; Allegra, C.J.; Johnston, P.G.; Kinsella, T.J. Cell cycle regulation of the G0/G1 transition in 5-fluorouracil-sensitive and -resistant human colon cancer cell lines. Cancer J 2000, 6, 234–242. [Google Scholar]
- Geller, J.I.; Szekely-Szucs, K.; Petak, I.; Doyle, B.; Houghton, J.A. P21Cip1 is a critical mediator of the cytotoxic action of thymidylate synthase inhibitors in colorectal carcinoma cells. Cancer Res 2004, 64, 6296–6303. [Google Scholar]
- Georges, S.A.; Biery, M.C.; Kim, S.Y.; Schelter, J.M.; Guo, J.; Chang, A.N.; Jackson, A.L.; Carleton, M.O.; Linsley, P.S.; Cleary, M.A.; et al. Coordinated regulation of cell cycle transcripts by p53-Inducible microRNAs, miR-192 and miR-215. Cancer Res 2008, 68, 10105–10112. [Google Scholar]
- Braun, C.J.; Zhang, X.; Savelyeva, I.; Wolff, S.; Moll, U.M.; Schepeler, T.; Ørntoft, T.F.; Andersen, C.L.; Dobbelstein, M. p53-Responsive micrornas 192 and 215 are capable of inducing cell cycle arrest. Cancer Res 2008, 68, 10094–10104. [Google Scholar]
- Schetter, A.J.; Leung, S.Y.; Sohn, J.J.; Zanetti, K.A.; Bowman, E.D.; Yanaihara, N.; Yuen, S.T.; Chan, T.L.; Kwong, D.L.; Au, G.K.; et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. J. Am. Med. Assoc 2008, 299, 425–436. [Google Scholar]
- Chen, X.; Guo, X.; Zhang, H.; Xiang, Y.; Chen, J.; Yin, Y.; Cai, X.; Wang, K.; Wang, G.; Ba, Y.; et al. Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene 2009, 28, 1385–1392. [Google Scholar]
- Kogo, R.; Shimamura, T.; Mimori, K.; Kawahara, K.; Imoto, S.; Sudo, T.; Tanaka, F.; Shibata, K.; Suzuki, A.; Komune, S.; et al. Long noncoding RNA HOTAIR regulates Polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res 2011, 71, 6320–6326. [Google Scholar]
- Blower, P.E.; Verducci, J.S.; Lin, S.; Zhou, J.; Chung, J.H.; Dai, Z.; Liu, C.G.; Reinhold, W.; Lorenzi, P.L.; Kaldjian, E.P.; et al. MicroRNA expression profiles for the NCI-60 cancer cell panel. Mol. Cancer Ther 2007, 6, 1483–1491. [Google Scholar]
- Blower, P.E.; Chung, J.H.; Verducci, J.S.; Lin, S.; Park, J.K.; Dai, Z.; Liu, C.G.; Schmittgen, T.D.; Reinhold, W.C.; Croce, C.M.; et al. MicroRNAs modulate the chemosensitivity of tumor cells. Mol. Cancer Ther 2008, 7, 1–9. [Google Scholar]
- Longley, D.B.; Allen, W.L.; Johnston, P.G. Drug resistance, predictive markers and pharmacogenomics in colorectal cancer. Biochim. Biophys. Acta 2006, 1766, 184–196. [Google Scholar]
- Szakacs, G.; Paterson, J.K.; Ludwig, J.A.; Booth-Genthe, C.; Gottesman, M.M. Targeting multidrug resistance in cancer. Nat. Rev. Drug Discov 2006, 5, 219–234. [Google Scholar]
- Iwasa, Y.; Nowak, M.A.; Michor, F. Evolution of resistance during clonal expansion. Genetics 2006, 172, 2557–2566. [Google Scholar]
- Roberti, A.; La Sala, D.; Cinti, C. Multiple genetic and epigenetic interacting mechanisms contribute to clonally selection of drug-resistant tumors: Current views and new therapeutic prospective. J. Cell. Physiol 2006, 207, 571–581. [Google Scholar]
- Glasspool, R.M.; Teodoridis, J.M.; Brown, R. Epigenetics as a mechanism driving polygenic clinical drug resistance. Br. J. Cancer 2006, 94, 1087–1092. [Google Scholar]
- Fojo, T. Multiple paths to a drug resistance phenotype: mutations, translocations, deletions and amplification of coding genes or promoter regions, epigenetic changes and microRNAs. Drug Resist. Updat 2007, 10, 59–67. [Google Scholar]
- Duesberg, P.; Li, R.; Sachs, R.; Fabarius, A.; Upender, M.B.; Hehlmann, R. Cancer drug resistance: the central role of the karyotype. Drug Resist. Updat 2007, 10, 51–58. [Google Scholar]
- Ragusa, M.; Majorana, A.; Statello, L.; Maugeri, M.; Salito, L.; Barbagallo, D.; Guglielmino, M.R.; Duro, L.R.; Angelica, R.; Caltabiano, R.; Biondi, A.; et al. Specific alterations of microRNA transcriptome and global network structure in colorectal carcinoma after cetuximab treatment. Mol. Cancer Ther 2010, 9, 3396–3409. [Google Scholar]
- Nakajima, G.; Hayashi, K.; Xi, Y.; Kudo, K.; Uchida, K.; Takasaki, K.; Yamamoto, M.; Ju, J. Non-coding MicroRNAs hsa-let-7g and hsa-miR-181b are associated with chemoresponse to S-1 in colon cancer. Cancer Genomics Proteomics 2006, 3, 317–3124. [Google Scholar]
- Fujita, Y.; Kojima, K.; Hamada, N.; Ohhashi, R.; Akao, Y.; Nozawa, Y.; Deguchi, T.; Ito, M. Effects of miR-34a on cell growth and chemoresistance in prostate cancer PC3 cells. Biochem. Biophys. Res. Commun 2008, 377, 114–119. [Google Scholar]
- Valeri, N.; Gasparini, P.; Braconi, C.; Paone, A.; Lovat, F.; Fabbri, M.; Sumani, K.M.; Alder, H.; Amadori, D.; Patel, T.; et al. MicroRNA-21 induces resistance to 5-fluorouracil by down-regulating human DNA MutS homolog 2 (hMSH2). Proc. Natl. Acad. Sci. USA 2010, 107, 21098–21103. [Google Scholar]
- Zhou, J.; Zhou, Y.; Yin, B.; Hao, W.; Zhao, L.; Ju, W.; Bai, C. 5-Fluorouracil and oxaliplatin modify the expression profiles of microRNAs in human colon cancer cells in vitro. Oncol. Rep 2010, 23, 121–128. [Google Scholar]
- Svoboda, M.; Izakovicova Holla, L.; Sefr, R.; Vrtkova, I.; Kocakova, I.; Tichy, B.; Dvorak, J. Micro-RNAs miR125b and miR137 are frequently upregulated in response to capecitabine chemoradiotherapy of rectal cancer. Int. J. Oncol 2008, 33, 541–547. [Google Scholar]
- Saunders, M.A.; Liang, H.; Li, W.H. Human polymorphism at microRNAs and microRNA target sites. Proc. Natl. Acad. Sci. USA 2007, 104, 3300–3305. [Google Scholar]
- Xing, J.; Wan, S.; Zhou, F.; Qu, F.; Li, B.; Myers, R.E.; Fu, X.; Palazzo, J.P.; He, X.; Chen, Z.; et al. Genetic polymorphisms in pre-microRNA genes as prognostic markers of colorectal cancer. Cancer Epidemiol. Biomarkers Prev 2011, in press. [Google Scholar]
- Boni, V.; Zarate, R.; Villa, J.C.; Bandrés, E.; Gomez, M.A.; Maiello, E.; Garcia-Foncillas, J.; Aranda, E. Role of primary miRNA polymorphic variants in metastatic colon cancer patients treated with 5-fluorouracil and irinotecan. Pharmacogenomics J 2010, 29, 1–8. [Google Scholar]
- Landi, D.; Gemignani, F.; Naccarati, A.; Pardini, B.; Vodicka, P.; Vodickova, L.; Novotny, J.; Forsti, A.; Hemminki, K.; Canzian, F. Polymorphisms within micro-RNA-binding sites and risk of sporadic colorectal cancer. Carcinogenesis 2008, 29, 579–584. [Google Scholar]
- Smits, K.M.; Paranjape, T.; Nallur, S.; Wouters, K.A.; Weijenberg, M.P.; Schouten, L.J.; van den Brandt, P.A.; Bosman, F.; Weidhaas, J.B.; van Engeland, M. A let-7 microRNA SNP in the KRAS 3′-UTR is prognostic in early-stage colorectal cancer. Clin. Cancer Res 2011, in press. [Google Scholar]
- Schepeler, T.; Reinert, J.T.; Ostenfeld, M.S.; Christensen, L.L.; Silahtaroglu, A.N.; Dyrskjøt, L.; Wiuf, C.; Sørensen, F.J.; Kruhøffer, M.; Laurberg, S.; et al. Diagnostic and prognostic microRNAs in stage II colon cancer. Cancer Res 2008, 68, 6416–6424. [Google Scholar]
- Wang, C.J.; Zhou, Z.G.; Wang, L.; Yang, L.; Zhou, B.; Gu, J.; Chen, H.Y.; Sun, X.F. Clinicopathological significance of microRNA-31, -143 and -145 expression in colorectal cancer. Dis. Markers 2009, 26, 27–34. [Google Scholar]
- Slaby, O.; Svoboda, M.; Fabian, P.; Smerdova, T.; Knoflickova, D.; Bednarikova, M.; Nenutil, R.; Vyzula, R. Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology 2007, 72, 397–402. [Google Scholar]
- Motoyama, K.; Inoue, H.; Takatsuno, Y.; Tanaka, F.; Mimori, K.; Uetake, H.; Sugihara, K.; Mori, M. Over- and under-expressed microRNAs in human colorectal cancer. Int. J. Oncol 2009, 34, 1069–1075. [Google Scholar]
- Lanza, G.; Ferracin, M.; Gafà, R.; Veronese, A.; Spizzo, R.; Pichiorri, F.; Liu, C.G.; Calin, G.A.; Croce, C.M.; Negrini, M. mRNA/microRNA gene expression profile in microsatellite unstable colorectal cancer. Mol. Cancer 2007, 6. [Google Scholar] [CrossRef]
- Wang, J.; Chen, J.; Chang, P.; LeBlanc, A.; Li, D.; Abbruzzesse, J.L.; Frazier, M.L.; Killary, A.M.; Sen, S. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev. Res. (Phila Pa) 2009, 2, 807–813. [Google Scholar]
- Van Deerlin, V.M.; Gill, L.H.; Nelson, P.T. Optimizing gene expression analysis in archival brain tissue. Neurochem. Res 2002, 27, 993–1003. [Google Scholar]
- Nelson, P.T.; Baldwin, D.A.; Scearce, L.M.; Oberholtzer, J.C.; Tobias, J.W.; Mourelatos, Z. Microarray-based, high-throughput gene expression profiling of microRNAs. Nat. Methods 2004, 1, 155–161. [Google Scholar]
- Li, J.; Smyth, P.; Flavin, R.; Cahill, S.; Denning, K.; Aherne, S.; Guenther, S.M.; O’Leary, J.J.; Sheils, O. Comparison of miRNA expression patterns using total RNA extracted from matched samples of formalin-fixed paraffin-embedded (FFPE) cells and snap frozen cells. BMC Biotechnol 2007, 7. [Google Scholar] [CrossRef]
- Ng, E.K.; Chong, W.W.; Jin, H.; Lam, E.K.; Shin, V.Y.; Yu, J.; Poon, T.C.; Ng, S.S.; Sung, J.J. Differential expression of microRNAs in plasma of patients with colorectal cancer: A potential marker for colorectal cancer screening. Gut 2009, 58, 1375–1381. [Google Scholar]
- Link, A.; Balaguer, F.; Shen, Y.; Nagasaka, T.; Lozano, J.J.; Boland, C.R.; Goel, A. Fecal MicroRNAs as novel biomarkers for colon cancer screening. Cancer Epidemiol. Biomarkers Prev 2010, 19, 1766–1774. [Google Scholar]
- Tsujiura, M.; Ichikawa, D.; Komatsu, S.; Shiozaki, A.; Takeshita, H.; Kosuga, T.; Konishi, H.; Morimura, R.; Deguchi, K.; Fujiwara, H. Circulating microRNAs in plasma of patients with gastric cancers. Br. J. Cancer 2010, 102, 1174–1179. [Google Scholar]
- Heneghan, H.M.; Miller, N.; Lowery, A.J.; Sweeney, K.J.; Newell, J.; Kerin, M.J. Circulating microRNAs as novel minimally invasive biomarkers for breast cancer. Ann. Surg 2010, 251, 499–505. [Google Scholar]
- Hu, Z.; Chen, X.; Zhao, Y.; Tian, T.; Jin, G.; Shu, Y.; Chen, Y.; Xu, L.; Zen, K.; Zhang, C. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J. Clin. Oncol 2010, 28, 1721–1726. [Google Scholar]
- Lawrie, C.H.; Gal, S.; Dunlop, H.M.; Pushkaran, B.; Liggins, A.P.; Pulford, K.; Banham, A.H.; Pezzella, F.; Boultwood, J.; Wainscoat, J.S. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br. J. Haematol 2008, 141, 672–675. [Google Scholar]
- Qu, H.; Xu, W.; Huang, Y.; Yang, S. Circulating miRNAs: Promising biomarkers of human cancer. Asian Pac. J. Cancer Prev 2011, 12, 1117–1125. [Google Scholar]
- Elmén, J.; Lindow, M.; Schütz, S.; Lawrence, M.; Petri, A.; Obad, S.; Lindholm, M.; Hedtjärn, M.; Hansen, H.F.; Berger, U.; et al. LNA-mediated microRNA silencing in non-human primates. Nature 2008, 452, 896–899. [Google Scholar]
- Krützfeldt, J.; Rajewsky, N.; Braich, R.; Rajeev, K.G.; Tuschl, T.; Manoharan, M.; Stoffel, M. Silencing of microRNAs in vivo with “antagomirs”. Nature 2005, 438, 685–689. [Google Scholar]
- Esau, C.; Davis, S.; Murray, S.F.; Yu, X.X.; Pandey, S.K.; Pear, M.; Watts, L.; Booten, S.L.; Graham, M.; McKay, R.; et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab 2006, 3, 87–98. [Google Scholar]
- Krichevsky, A.M.; Gabriely, G. miR-21: A small multi-faceted RNA. J. Cell. Mol. Med 2009, 13, 39–53. [Google Scholar]
- Si, M.L.; Zhu, S.; Wu, H.; Lu, Z.; Wu, F.; Mo, Y.Y. miR-21-mediated tumor growth. Oncogene 2007, 26, 2799–2803. [Google Scholar]
- Gumireddy, K.; Young, D.D.; Xiong, X.; Hogenesch, J.B.; Huang, Q.; Deiters, A. Small-molecule inhibitors of microrna miR-21 function. Angew. Chem. Int. Ed. Engl 2008, 47, 7482–7484. [Google Scholar]
- Ebert, M.S.; Neilson, J.R.; Sharp, P.A. MicroRNA sponges: Competitive inhibitors of small RNAs in mammalian cells. Nat. Methods 2007, 4, 721–726. [Google Scholar]
- Xiao, J.; Yang, B.; Lin, H.; Lu, Y.; Luo, X.; Wang, Z. Novel approaches for gene-specific interference via manipulating actions of microRNAs: Examination on the pacemaker channel genes HCN2 and HCN4. J. Cell. Physiol 2007, 212, 285–292. [Google Scholar]
- Ji, Q.; Hao, X.; Meng, Y.; Zhang, M.; Desano, J.; Fan, D.; Xu, L. Restoration of tumor suppressor miR-34 inhibits human p53-mutant gastric cancer tumorspheres. BMC Cancer 2008, 8. [Google Scholar] [CrossRef]
- Ji, Q.; Hao, X.; Zhang, M.; Tang, W.; Yang, M.; Li, L.; Xiang, D.; Desano, J.T.; Bommer, G.T.; Fan, D.; et al. MicroRNA miR-34 inhibits human pancreatic cancer tumor-initiating cells. PLoS One 2009, 4, e6816. [Google Scholar]
- Pirollo, K.F.; Xu, L.; Chang, E.H. Non-viral gene delivery for p53. Curr. Opin. Mol. Ther 2000, 2, 168–175. [Google Scholar]
- Xu, L.; Pirollo, K.F.; Chang, E.H. Tumor-targeted p53-gene therapy enhances the efficacy of conventional chemo/radiotherapy. J. Control. Release 2001, 74, 115–128. [Google Scholar]
- Ibrahim, A.F.; Weirauch, U.; Thomas, M.; Grunweller, A.; Hartmann, R.K.; Aigner, A. MiRNA replacement therapy through PEI-mediated in vivo delivery of miR-145 or miR-33a in colon carcinoma. Cancer Res 2011, 71, 5214–5224. [Google Scholar]
- Trang, P.; Wiggins, J.F.; Daige, C.L.; Cho, C.; Omotola, M.; Brown, D.; Weidhaas, J.B.; Bader, A.G.; Slack, F.J. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol. Ther 2011, 19, 1116–1122. [Google Scholar]
- Landen, C.N., Jr; Chavez-Reyes, A.; Bucana, C.; Schmandt, R.; Deavers, M.T.; Lopez-Berestein, G.; Sood, A.K. Therapeutic EphA2 gene targeting in vivo using neutral liposomal small interfering RNA delivery. Cancer Res 2005, 65, 6910–6918. [Google Scholar]
- Takeshita, F.; Patrawala, L.; Osaki, M.; Takahashi, R.U.; Yamamoto, Y.; Kosaka, N.; Kawamata, M.; Kelnar, K.; Bader, A.G.; Brown, D.; et al. Systemic delivery of synthetic microRNA-16 inhibits the growth of metastatic prostate tumors via downregulation of multiple cell-cycle genes. Mol. Ther 2010, 18, 181–187. [Google Scholar]
- Bonci, D.; Coppola, V.; Musumeci, M.; Addario, A.; Giuffrida, R.; Memeo, L.; D’Urso, L.; Pagliuca, A.; Biffoni, M.; Labbaye, C.; et al. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat. Med 2008, 14, 1271–1277. [Google Scholar]
- Liu, C.; Kelnar, K.; Liu, B.; Chen, X.; Calhoun-Davis, T.; Li, H.; Patrawala, L.; Yan, H.; Jeter, C.; Honorio, S.; et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat. Med 2011, 17, 211–215. [Google Scholar]
- Cho, W.C. Molecular diagnostics for monitoring and predicting therapeutic effect in cancer. Expert Rev. Mol. Diagn 2011, 11, 9–12. [Google Scholar]
- Zhang, W.; Dahlberg, J.E.; Tam, W. MiRNAs in tumorigenesis: A primer. Am. J. Pathol 2007, 171, 728–738. [Google Scholar]
- Henry, J.C.; Azevedo-Pouly, A.C.; Schmittgen, T.D. microRNA replacement therapy for cancer. Pharm. Res 2011, 28, 3030–3042. [Google Scholar]
| miRNA | Dysregulation | Clinical-related phenotypes | Reference |
|---|---|---|---|
| Let-7g | Upregulated in CRC | Higher level associated with poor S-1 response | [93] |
| miR-18a | Upregulated in CRC | Higher level associated with poor overall survival | [106] |
| miR-21 | Upregulated in adenoma, CRC, and liver metastasis tissue | Higher level associated with lymph node positivity, metastasis; poor survival, poor therapeutic outcomes, rapid recurrence; shorter disease-free interval | [80,105] |
| miR-31 | Upregulated in CRC | Higher level associated with higher TNM stages and local invasion | [61,105,108] |
| miR-106a | Upregulated in colon cancer | Higher level associated with longer disease-free survival and overall survival | [80] |
| miR-143 | Downregulated in colon cancer and liver metastasis tissue | Lower level associated with larger tumor size and longer disease-free interval | [105,106,108] |
| miR-145 | Downregulated in CRC | Lower level associated with larger tumor size; related with tumor location | [105,106,108] |
| miR-181b | Upregulated in CRC | Higher level associated with poor S-1 response | [93] |
| miR-320 | Downregulated in MSS tumor | Lower level associated with shorter progression-free survival | [103] |
| miR-498 | Downregulated in MSS tumor | Lower level associated with shorter progression-free survival | [103] |
© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Zarate, R.; Boni, V.; Bandres, E.; Garcia-Foncillas, J. MiRNAs and LincRNAs: Could They Be Considered as Biomarkers in Colorectal Cancer? Int. J. Mol. Sci. 2012, 13, 840-865. https://doi.org/10.3390/ijms13010840
Zarate R, Boni V, Bandres E, Garcia-Foncillas J. MiRNAs and LincRNAs: Could They Be Considered as Biomarkers in Colorectal Cancer? International Journal of Molecular Sciences. 2012; 13(1):840-865. https://doi.org/10.3390/ijms13010840
Chicago/Turabian StyleZarate, Ruth, Valentina Boni, Eva Bandres, and Jesús Garcia-Foncillas. 2012. "MiRNAs and LincRNAs: Could They Be Considered as Biomarkers in Colorectal Cancer?" International Journal of Molecular Sciences 13, no. 1: 840-865. https://doi.org/10.3390/ijms13010840




