Molecular Cloning and Functional Characterization of Tibetan Porcine STING
Abstract
:1. Introduction
2. Results and Discussion
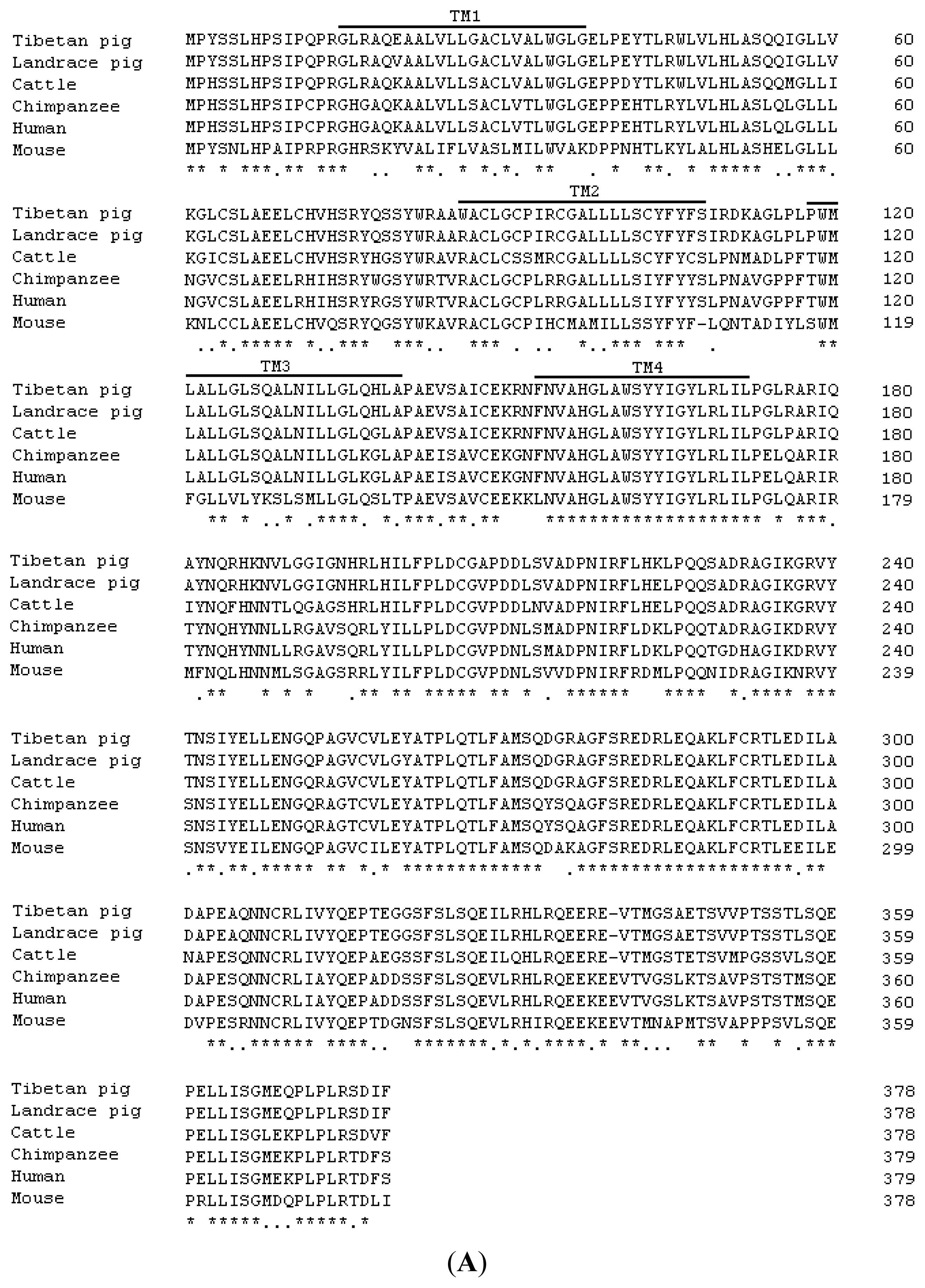
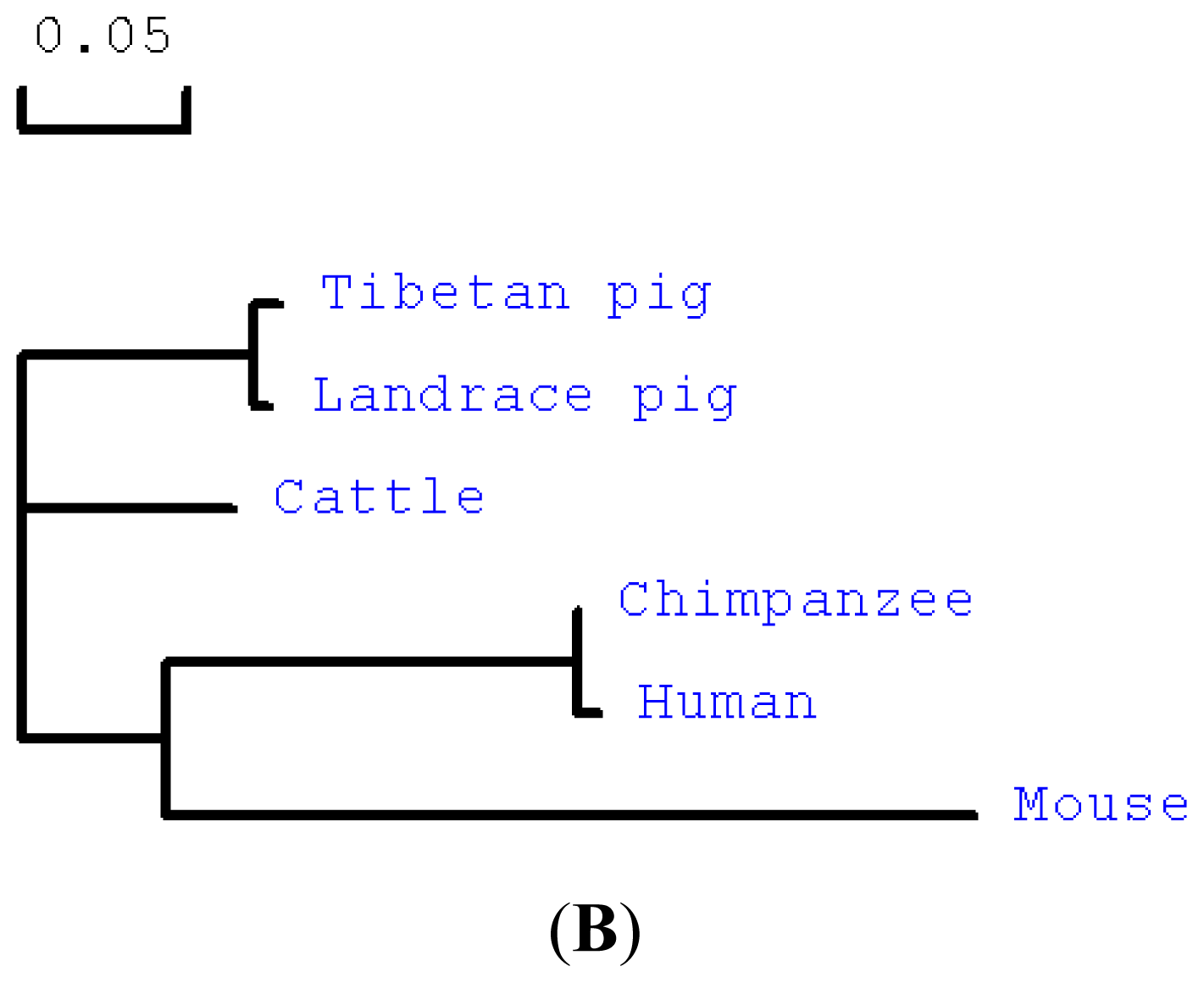
2.1. Cloning and Sequence Analysis of Tibetan Porcine STING
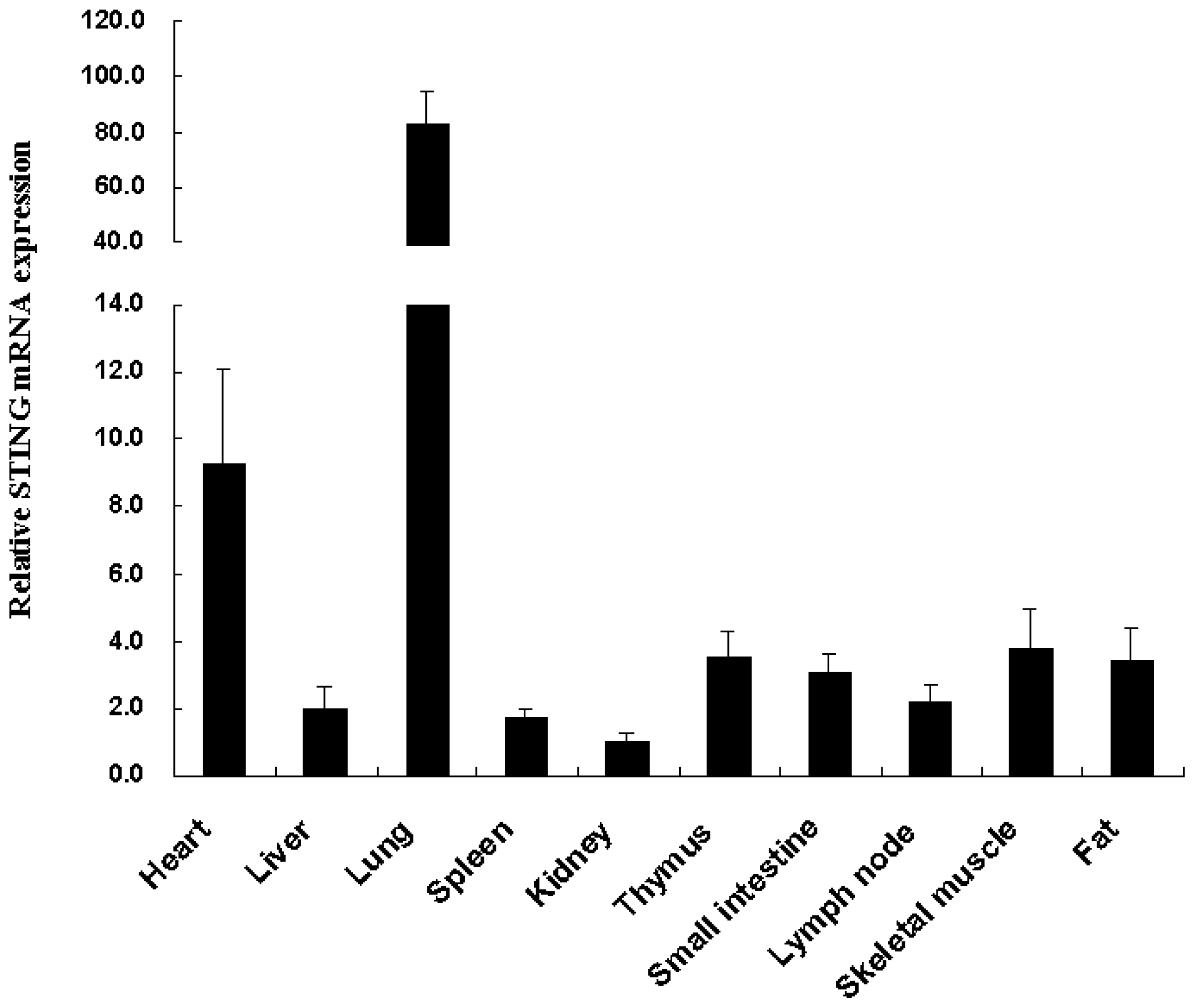
2.2. Tissue Distribution of the Tibetan Porcine STING mRNA
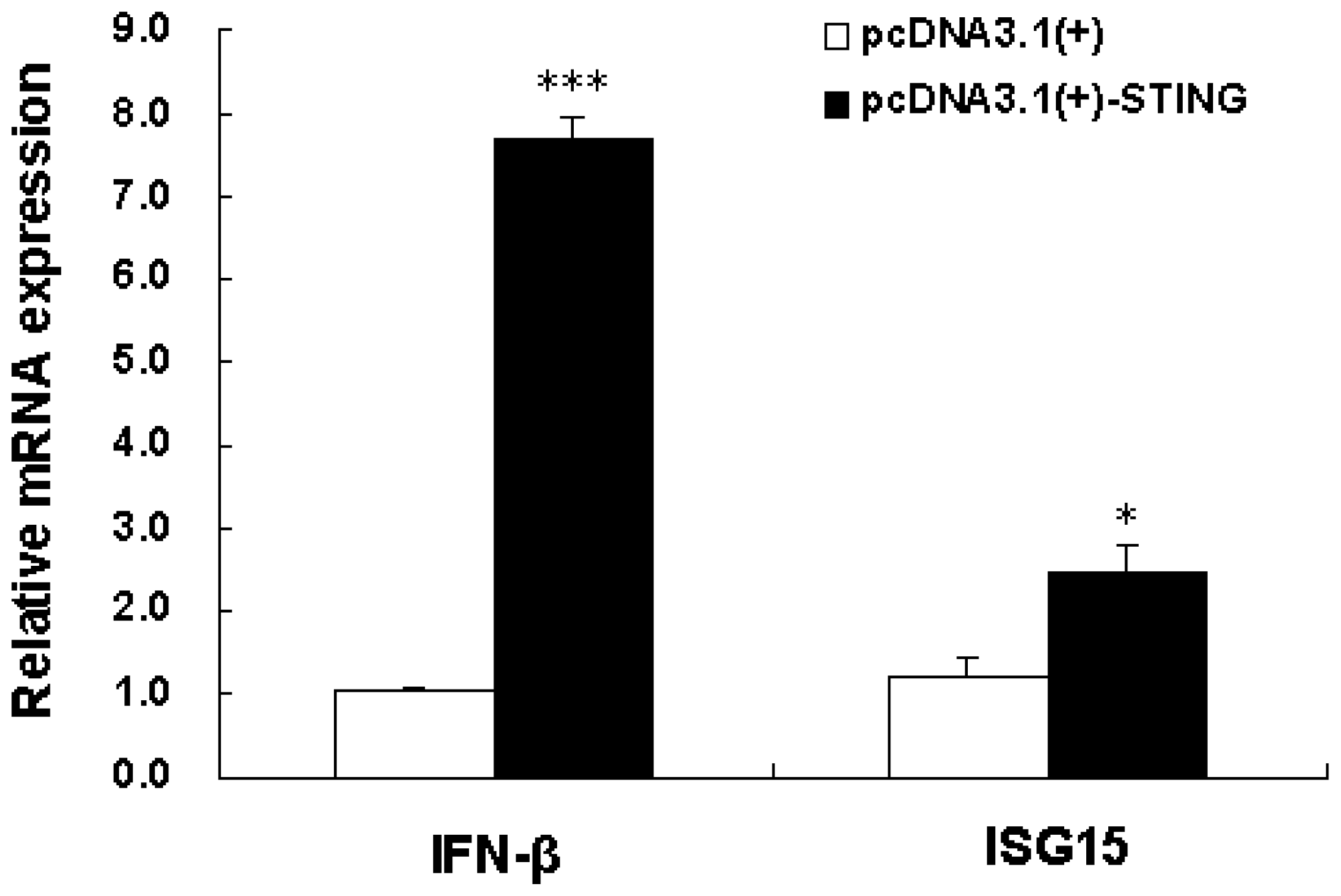
2.3. Overexpression of Tibetan Porcine STING Leads to Upregulation of IFN-β and ISG15 in IPEC-J2 Cells
3. Materials and Methods
3.1. Animals and Tissue Sample Collection
3.2. RNA Isolation and Reverse Transcription
3.3. Cloning of STING cDNA
3.4. Plasmid Construction
3.5. Cell Culture
3.6. Transfection
3.7. Real-Time Quantitative PCR
3.8. Statistical Analysis
4. Conclusions
Supplementary Material
ijms-13-00506-s001.pdfAcknowledgments
- Conflict of InterestThe authors declare no conflict of interest.
References
- Kawai, T.; Akira, S. The roles of TLRs, RLRs and NLRs in pathogen recognition. Int. Immunol 2009, 21, 317–337. [Google Scholar]
- Palm, N.W.; Medzhitov, R. Pattern recognition receptors and control of adaptive immunity. Immunol. Rev 2009, 227, 221–233. [Google Scholar]
- Takeuchi, O.; Akira, S. Innate immunity to virus infection. Immunol. Rev 2009, 227, 75–86. [Google Scholar]
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen recognition and innate immunity. Cell 2006, 124, 783–801. [Google Scholar]
- Takeuchi, O.; Akira, S. Recognition of viruses by innate immunity. Immunol. Rev 2007, 220, 214–224. [Google Scholar]
- Alexopoulou, L.; Holt, A.C.; Medzhitov, R.; Flavell, R.A. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 2001, 413, 732–738. [Google Scholar]
- Yoneyama, M.; Kikuchi, M.; Natsukawa, T.; Shinobu, N.; Imaizumi, T.; Miyagishi, M.; Taira, K.; Akira, S.; Fujita, T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol 2004, 5, 730–737. [Google Scholar]
- Yoneyama, M.; Kikuchi, M.; Matsumoto, K.; Imaizumi, T.; Miyagishi, M.; Taira, K.; Foy, E.; Loo, Y.M.; Gale, M., Jr; Akira, S.; Yonehara, S.; Kato, A.; Fujita, T. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol 2005, 175, 2851–2858. [Google Scholar]
- Nakhaei, P.; Hiscott, J.; Lin, R. STING-ing the antiviral pathway. J. Mol. Cell Biol 2010, 2, 110–112. [Google Scholar]
- Zeng, W.; Chen, Z.J. MITAgating viral infection. Immunity 2008, 29, 513–515. [Google Scholar]
- Ishikawa, H.; Barber, G.N. STING is an endoplasmic reticulum adaptor that facilitates innate immune signaling. Nature 2008, 455, 674–678. [Google Scholar]
- Sun, W.; Li, Y.; Chen, L.; Chen, H.; You, F.; Zhou, X.; Zhou, Y.; Zhai, Z.; Chen, D.; Jiang, Z. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc. Natl. Acad. Sci. USA 2009, 106, 8653–8658. [Google Scholar]
- Zhong, B.; Yang, Y.; Li, S.; Wang, Y.Y.; Li, Y.; Diao, F.; Lei, C.; He, X.; Zhang, L.; Tien, P.; Shu, H.B. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 2008, 29, 538–550. [Google Scholar]
- Xie, L.; Liu, M.; Fang, L.; Su, X.; Cai, K.; Wang, D.; Chen, H.; Xiao, S. Molecular cloning and functional characterization of porcine stimulator of interferon genes (STING). Dev. Comp. Immunol 2010, 34, 847–854. [Google Scholar]
- Bäckhed, F.; Ley, R.E.; Sonnenburg, J.L.; Peterson, D.A.; Gordon, J.I. Host-bacterial mutualism in the human intestine. Science 2005, 307, 1915–1920. [Google Scholar]
- Shao, L.; Serrano, D.; Mayer, L. The role of epithelial cells in immune regulation in the gut. Semin. Immunol 2001, 13, 163–175. [Google Scholar]
- Martensen, P.M.; Justesen, J. Small ISGs coming forward. J. Interferon Cytokine Res 2004, 24, 1–19. [Google Scholar]
- Rhoads, J.M.; Chen, W.; Chu, P.; Berschneider, H.M.; Argenzio, R.A.; Paradiso, A.M. L-Glutamine and L-asparagine stimulate Na+-H+ exchange in porcine jejunal enterocytes. Am. J. Physiol 1994, 266, G828–G838. [Google Scholar]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001, 29. [Google Scholar] [CrossRef]
| Gene | Primer | Sequence | GenBank ID | Product size |
|---|---|---|---|---|
| IFN-β | Forward | 5′-AAATCGCTCTCCTGATGTGT-3′ | NM_001003923 | 78 bp |
| Reverse | 5′-TGCTCCTTTGTTGGTATCG-3′ | |||
| ISG15 | Forward | 5′-AGCAACGCCTATGAGGTC-3′ | EU584557 | 101 bp |
| Reverse | 5′-AAAGTCAGCCAGAAATGGTC-3′ | |||
| β-actin | Forward | 5′-CCACGAAACTACCTTCAACTCC-3′ | DQ845171 | 132 bp |
| Reverse | 5′-GTGATCTCCTTCTGCATCCTGT-3′ |
© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Huang, Z.; Chen, X.; Zhang, K.; Yu, B.; Mao, X.; Zhao, Y.; Chen, D. Molecular Cloning and Functional Characterization of Tibetan Porcine STING. Int. J. Mol. Sci. 2012, 13, 506-515. https://doi.org/10.3390/ijms13010506
Huang Z, Chen X, Zhang K, Yu B, Mao X, Zhao Y, Chen D. Molecular Cloning and Functional Characterization of Tibetan Porcine STING. International Journal of Molecular Sciences. 2012; 13(1):506-515. https://doi.org/10.3390/ijms13010506
Chicago/Turabian StyleHuang, Zhiqing, Xiaoling Chen, Keying Zhang, Bing Yu, Xiangbing Mao, Ye Zhao, and Daiwen Chen. 2012. "Molecular Cloning and Functional Characterization of Tibetan Porcine STING" International Journal of Molecular Sciences 13, no. 1: 506-515. https://doi.org/10.3390/ijms13010506




