The Effect of Secretory Factors of Adipose-Derived Stem Cells on Human Keratinocytes
Abstract
:1. Introduction
2. Results and Discussion
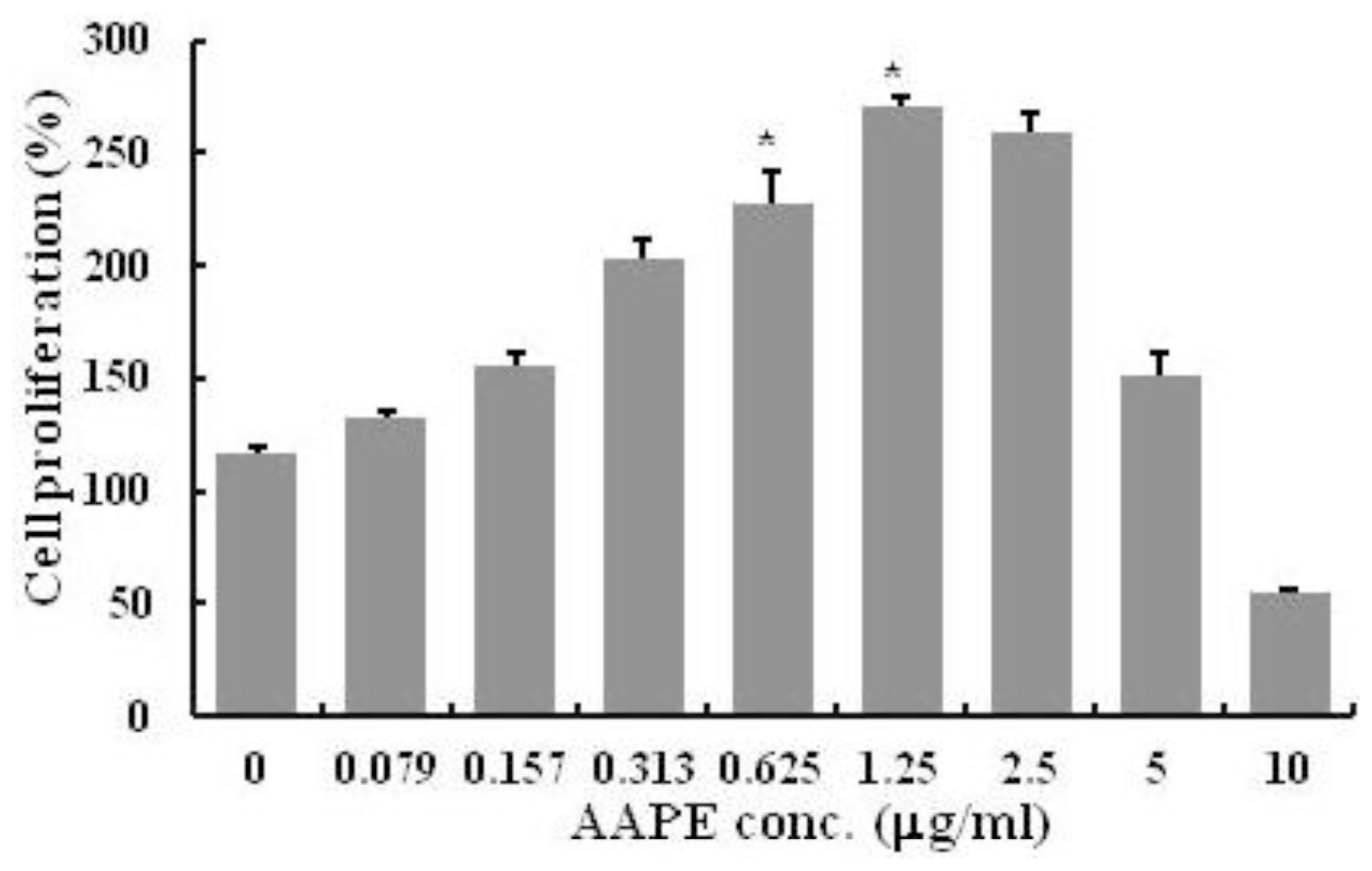
2.1. HK Proliferation
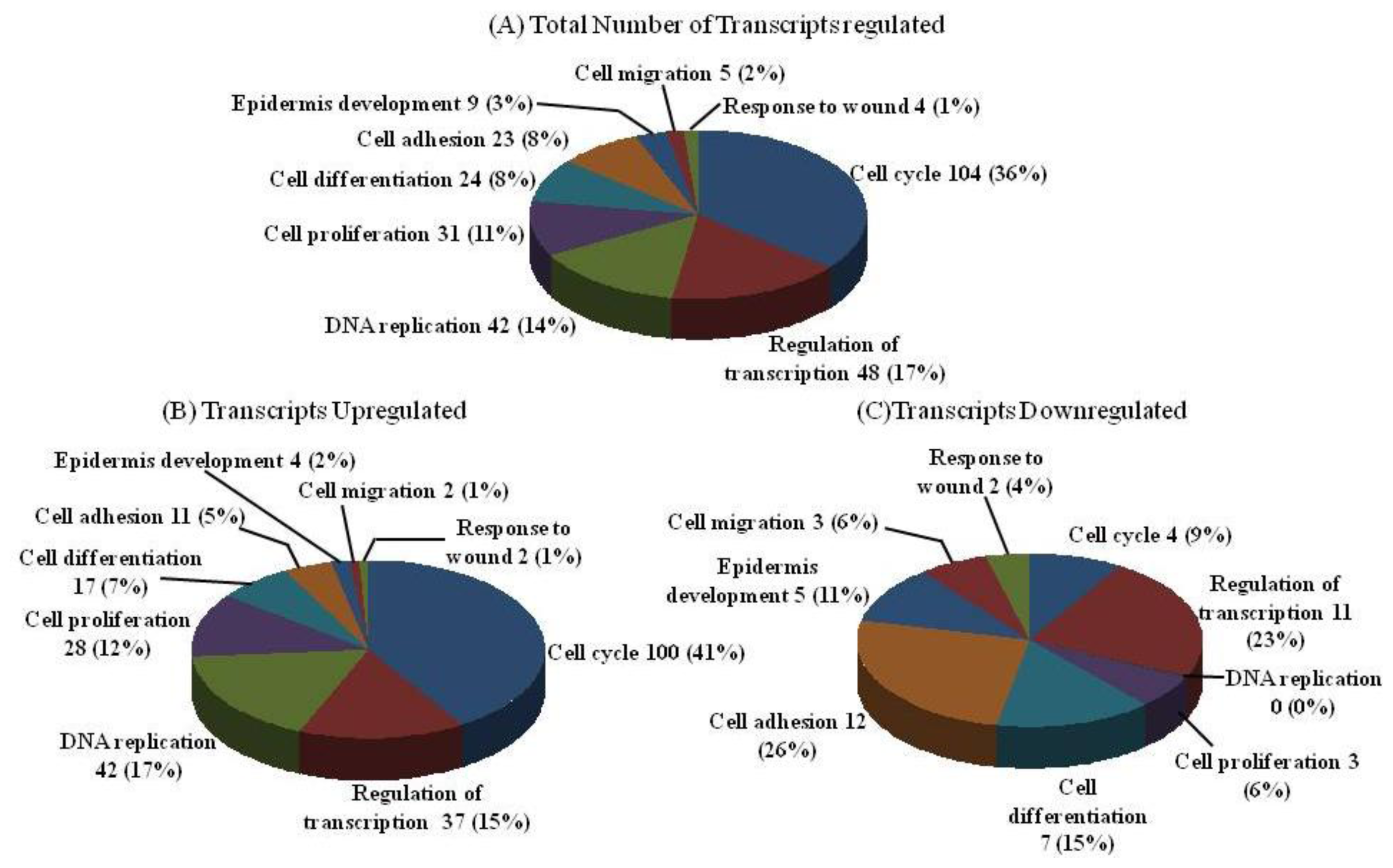
2.2. DNA Chip Analysis
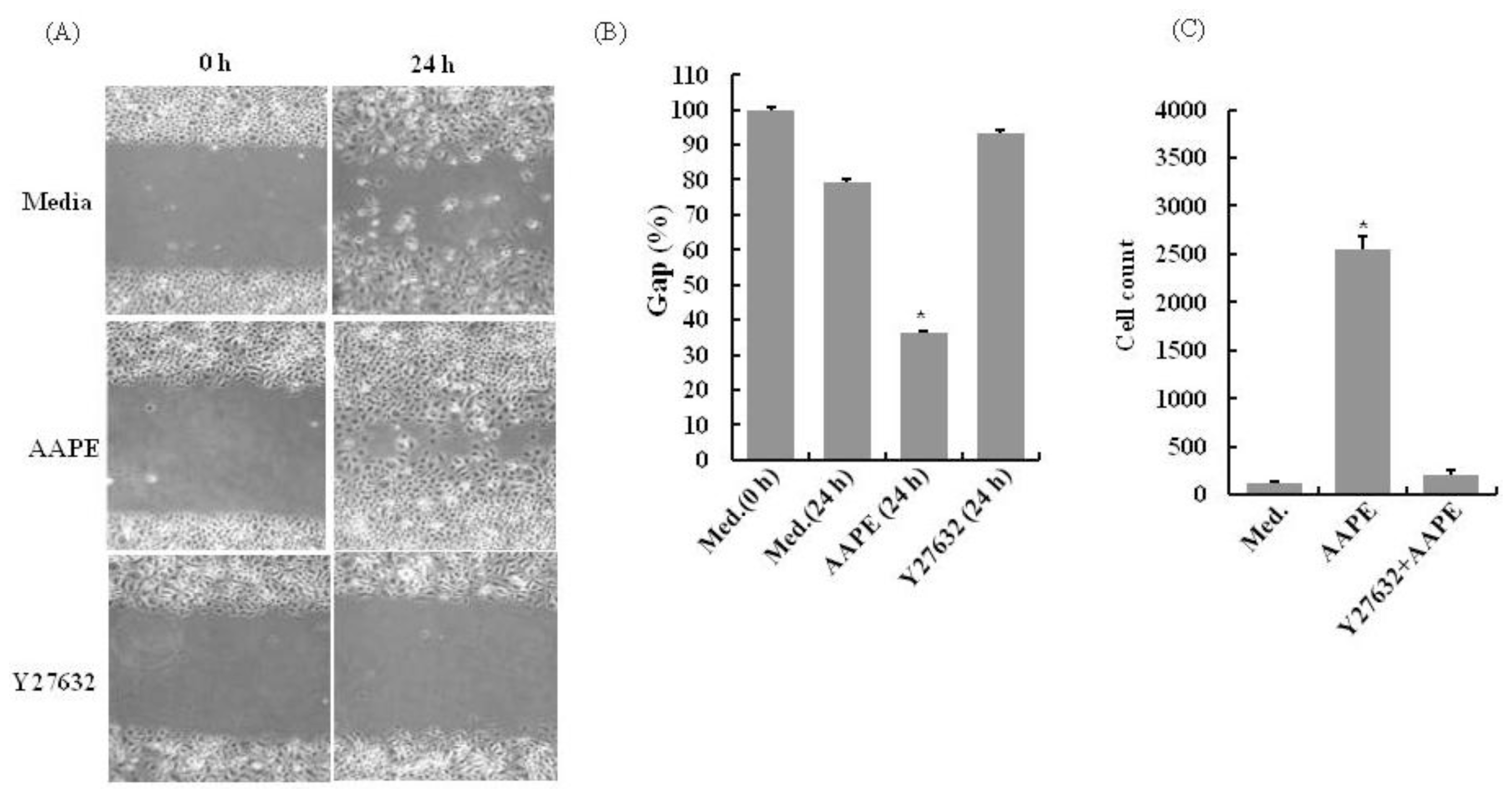
2.3. AAPE Stimulates Wounding Healing Cell Migration via ROCK Pathway
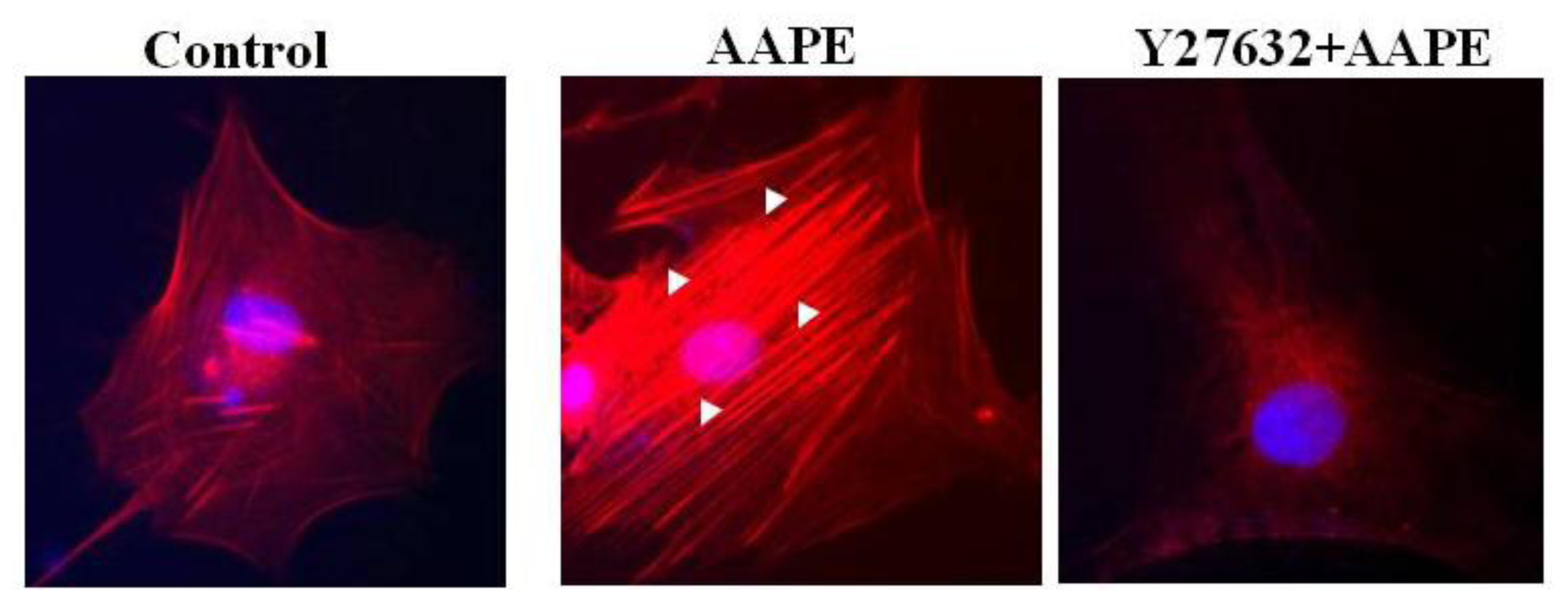
2.4. AAPE Augments Stress Fiber Formation in HK
2.5. RhoA-ROCK Pathway Is Involved in Actin Stress Fiber Formation in HK
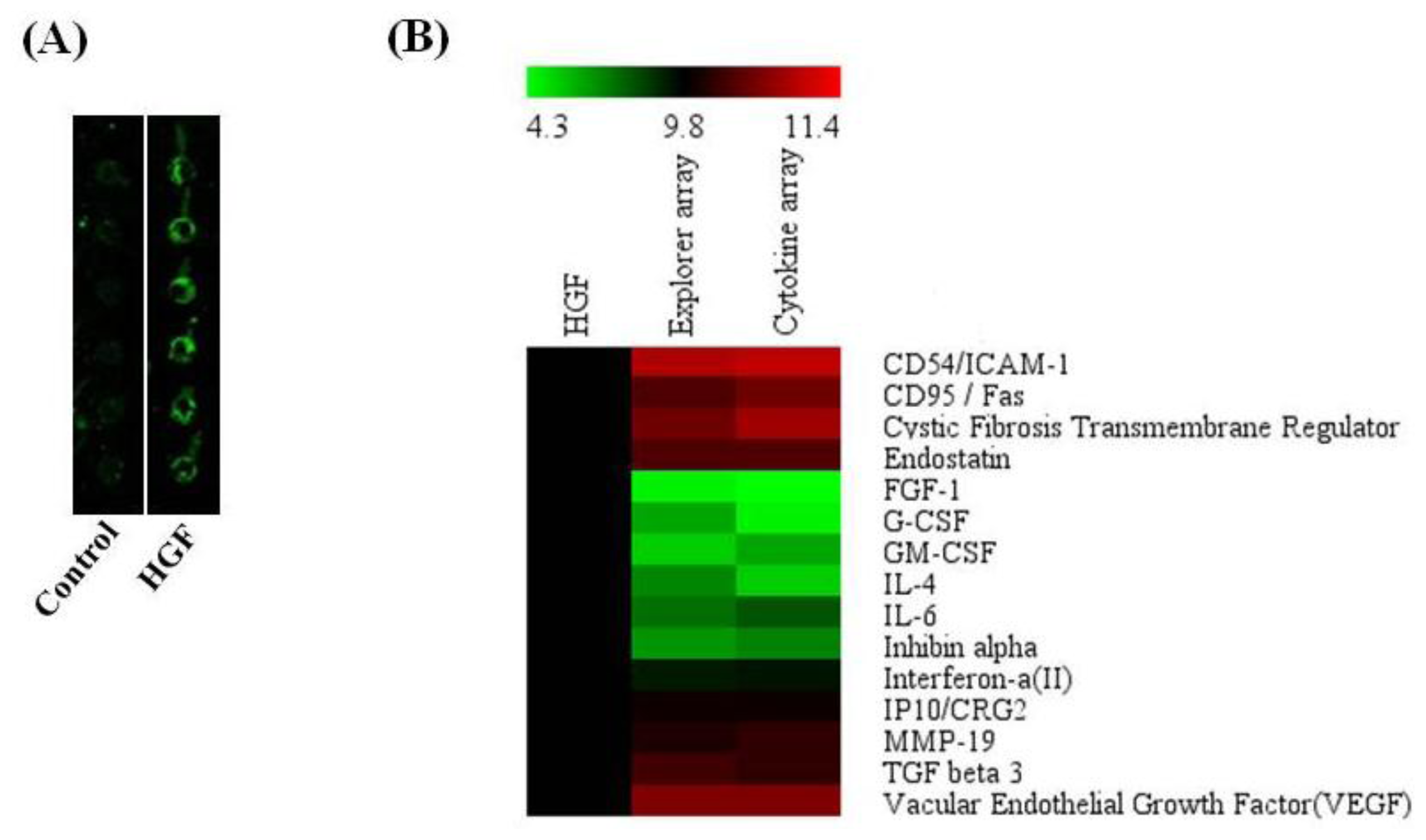
2.7. Antibody Array
2.8. Discussion
3. Materials and Methods
3.1. Stem Cell Culture and AAPE Production
3.2. Cell Culture
3.3. Cell Proliferation
3.4. DNA Chip Analysis
3.5. In vitro Migration Assay
3.6. Chemotaxis Assay
3.7. Fluorescence Microscopy for Stress Fiber Formation
3.8. RhoA Pull Down
3.9. Western Blot
3.10. Proteome Analysis
3.11. Antibody Array
4. Conclusions
Acknowledgements
References
- Sterodimas, A.; De Faria, J.; Nicaretta, B.; Pitanguy, I. Tissue engineering with adipose-derived stem cells (ADSCs): Current and future applications. J. Plast. Reconstr. Aesthet. Surg 2010, 63, 1886–1892. [Google Scholar]
- Moyer, H.R.; Kinney, R.C.; Singh, K.A.; Williams, J.K.; Schwartz, Z.; Boyan, B.D. Alginate microencapsulation technology for the percutaneous delivery of adipose-derived stem cells. Ann. Plast. Surg 2010, 65, 497–503. [Google Scholar]
- Kim, W.S.; Park, B.S.; Sung, J.H. Protective role of adipose-derived stem cells and their soluble factors in photoaging. Arch. Dermatol. Res 2009, 301, 329–336. [Google Scholar]
- Kim, W.S.; Park, S.H.; Ahn, S.J.; Kim, H.K.; Park, J.S.; Lee, G.Y.; Kim, K.J.; Whang, K.K.; Kang, S.H.; Park, B.S.; et al. Whitening effect of adipose-derived stem cells: A critical role of TGF-beta 1. Biol. Pharm. Bull 2008, 31, 606–610. [Google Scholar]
- Wright, C.S.; Van Steensel, M.A.; Hodgins, M.B.; Martin, P.E. Connexin mimetic peptidesimprove cell migration rates of human epidermal keratinocytes and dermal fibroblasts in vitro. Wound Repair Regen 2009, 17, 240–249. [Google Scholar]
- Mansbridge, J. Skin tissue engineering. J. Biomater. Sci. Polym. Ed 2008, 19, 955–968. [Google Scholar]
- Nolte, S.V.; Xu, W.; Rennekampff, H.O.; Rodemann, H.P. Diversity of fibroblasts a review on implications for skin tissue engineering. Cells Tissues Organs 2008, 187, 165–176. [Google Scholar]
- Xia, W.; Phan, T.T.; Lim, I.J.; Longaker, M.T.; Yang, G.P. Complex epithelial-mesenchymal interactions modulate transforming growth factor-beta expression in keloid-derived cells. Wound Repair Regen 2004, 12, 546–556. [Google Scholar]
- Mansbridge, J. Skin tissue engineering. J. Biomater. Sci. Polym. Ed 2008, 19, 955–968. [Google Scholar]
- Nobes, C.D.; Hall, A. Rho GTPases control polarity, protrusion, and adhesion during cell movement. J. Cell Biol 1999, 144, 1235–1244. [Google Scholar]
- Cramer, L.P.; Siebert, M.; Mitchison, T.J. Identification of novel graded polarity actin filament Bundles in locomoting heart fibroblasts:implications for the generation of motile force. J. Cell Biol 1997, 136, 1287–1305. [Google Scholar]
- Adams, J.C. Formation of stable microspikes containing actin and the 55 kDa actin bundling protein actin and the 55 kDa actin bundling protein, fascin, is a consequence of cell adhesion to thrombospondin-1: Implications for the anti-adhesive activities of thrombospondin-1. J. Cell Sci 1995, 108, 1977–1990. [Google Scholar]
- Chen, B.; Li, A.; Wang, D.; Wang, M.; Zheng, L.; Bartles, J.R. Espin contains an additional actin-binding site in its N terminus and is a major actin-bundling protein of the sertoli cell-spermatid ectoplasmic specialization junctional plaque. Mol. Biol. Cell 1999, 10, 4327–4339. [Google Scholar]
- Wang, K.; Ash, J.F.; Singer, S.J. Filamin, a new high-molecular-weight protein found in smooth muscle and non-muscle cells. Proc. Natl. Acad. Sci. USA 1975, 72, 4483–4486. [Google Scholar]
- Zhang, L.; Deng, M.; Parthasarathy, R.; Wang, L.; Mongan, M.; Molkentin, J.D.; Zheng, Y.; Xia, Y. MEKK1 transduces activin signals in keratinocytes to induce an actin stress fiber formation and migration. Mol. Cell Biol 2005, 25, 60–65. [Google Scholar]
- Rid, R.; Schiefermeier, N.; Grigoriev, I.; Small, J.V.; Kaverina, I. The last but not the least: The origin and significance of trailing adhesions in fibroblastic cells. Cell Motil. Cytoskeleton 2005, 61, 161–171. [Google Scholar]
- Xu, D.; Kishi, H.; Kawamichi, H.; Kajiya, K.; Takada, Y.; Kobayashi, S. Sphingosylphosphorylcholine induces stress fiber formation via activation of Fyn-RhoA-ROCK signaling pathway in fibroblasts. Cell Signal 2012, 24, 282–289. [Google Scholar]
- Jackson, B.; Peyrollier, K.; Pedersen, E.; Basse, A.; Karlsson, R.; Wang, Z.; Lefever, T.; Ochsenbein, A.M.; Schmidt, G.; Aktories, K.; et al. RhoA is dispensable for skin development, but crucial for contraction and directed migration of keratinocytes. Mol. Biol. Cell 2011, 22, 593–605. [Google Scholar]
- Igarashi, A.; Okochi, H.; Bradham, D.M.; Grotendorst, G.R. Regulation of connective tissue growth factor gene expression in human skin fibroblasts and during wound repair. Mol. Biol. Cell 1993, 4, 637–645. [Google Scholar]
- Sato, M.; Sawamura, D.; Ina, S.; Yaguchi, T.; Hanada, K.; Hashimoto, I. In vivo introduction of the interleukin 6 gene into human keratinocytes: Induction of epidermal proliferation by the fully spliced form of interleukin 6, but not by the alternatively spliced form. Arch. Dermatol. Res 1999, 291, 400–404. [Google Scholar]
- Peschen, M.; Grenz, H.; Brand-Saberi, B.; Bunaes, M.; Simon, J.C.; Schöpf, E.; Vanscheidt, W. Increased expression of platelet-derived growth factor receptor alpha and beta and vascular endothelial growth factor in the skin of patients with chronic venous insufficiency. Arch. Dermatol. Res 1998, 290, 291–297. [Google Scholar]
- Gallucci, R.M.; Sloan, D.K.; Heck, J.M.; Murray, A.R.; O’Dell, S.J. Interleukin 6 indirectly induces keratinocyte migration. J. Invest. Dermatol 2004, 122, 764–772. [Google Scholar]
- Peura, M.; Bizik, J.; Salmenperä, P.; Noro, A.; Korhonen, M.; Pätilä, T.; Vento, A.; Vaheri, A.; Alitalo, R.; Vuola, J.; et al. Bone marrow mesenchymal stem cells undergo nemosis and induce keratinocyte wound healing utilizing the HGF/c-Met/PI3K pathway. Wound Repair Regen 2009, 17, 569–577. [Google Scholar]
- Mann, A.; Breuhahn, K.; Schirmacher, P.; Blessing, M. Keratinocyte-derived granulocyte-macrophage colony stimulating factor accelerates wound healing: Stimulation of keratinocyte proliferation, granulation tissue formation, and vascularization. J. Invest. Dermatol 2001, 117, 1382–1390. [Google Scholar]
- Ren, H.; Cao, Y.; Zhao, Q.; Li, J.; Zhou, C.; Liao, L.; Jia, M.; Zhao, Q.; Cai, H.; Han, Z.C.; et al. Proliferation and differentiation of bone marrow stromal cells under hypoxic conditions. Biochem. Biophys. Res. Commun 2006, 347, 12–21. [Google Scholar]
- Efimenko, AIu.; Starostina, E.E.; Rubina, K.A.; Kalinina, N.I.; Parfenova, E.V. Viability and angiogenic activity of mesenchymal stromal cells from adipose tissue and bone marrow in hypoxia and inflammation in vitro. Tsitologiia 2010, 52, 144–154. [Google Scholar]
- Martin, P. Wound healing-aiming for perfect skin regeneration. Science 1997, 276, 75–81. [Google Scholar]
- Singer, A.J.; Clark, R.A. Cutaneous wound healing. N. Engl. J. Med 1999, 341, 738–746. [Google Scholar]
- Shephard, P.; Martin, G.; Smola-Hess, S.; Brunner, G.; Krieg, T.; Smola, H. Myofibroblast differentiation is induced in keratinocyte-fibroblast co-cultures and is antagonistically regulated by endogenous transforming growth factor beta and interleukin-1. Am. J. Pathol 2004, 164, 2055–2066. [Google Scholar]
- Werner, S.; Krieg, T.; Smola, H. Keratinocyte-fibroblast interactions in wound healing. J. Invest. Dermatol 2007, 127, 998–1008. [Google Scholar]
- Ridley, A.J.; Schwartz, M.A.; Burridge, K.; Firtel, R.A.; Ginsberg, M.H.; Borisy, G.; Parsons, J.T.; Horwitz, A.R. Cell migration: Integrating signals from front to back. Science 2003, 302, 1704–1709. [Google Scholar]
- Bevan, D.; Gherardi, E.; Fan, T.P.; Edwards, D.; Warn, R. Diverse and potent activities of HGF/SF in skin wound repair. J. Pathol 2004, 203, 831–838. [Google Scholar]
- Dong, G.; Lee, T.L.; Yeh, N.T.; Geoghegan, J.; Van Waes, C.; Chen, Z. Metastatic squamous cell carcinoma cells that overexpress c-Met exhibit enhanced angiogenesis factor expression, scattering and metastasis in response to hepatocyte growth factor. Oncogene 2004, 23, 6199–6208. [Google Scholar]
- Delehedde, M.; Lyon, M.; Vidyasagar, R.; McDonnell, T.J.; Fernig, D.G. Hepatocyte growth factor/scatter factor binds to small heparin-derived oligosaccharides and stimulates the proliferation of human HaCaT keratinocytes. J. Biol. Chem 2002, 277, 12456–12462. [Google Scholar]
- Hinitt, C.A.; Wood, J.; Lee, S.S.; Williams, A.C.; Howarth, J.L.; Glover, C.P.; Uney, J.B.; Hague, A. BAG-1 enhances cell–cell adhesion, reduces proliferation and induces chaperone-independent suppression of hepatocyte growth factor-induced epidermal keratinocyte migration. Exp. Cell Res 2010, 316, 2042–2060. [Google Scholar]
- Huber, K. Plasminogen activator inhibitor type-1 (part one): Basic mechanisms, regulation, and role for thromboembolic disease. J. Thromb. Thrombolys 2001, 11, 183–193. [Google Scholar]
- Rømer, J.; Lund, L.R.; Eriksen, J.; Ralfkiaer, E.; Zeheb, R.; Gelehrter, T.D.; Danø, K.; Kristensen, P. Differential expression of urokinase-type plasminogen activator and its type-1 inhibitor during healing of mouse skin wounds. J. Invest. Dermatol 1991, 97, 803–811. [Google Scholar]
- Jensen, P.J.; Lavker, R.M. Modulation of the plasminogen activator cascade during enhanced epidermal proliferation in vivo. Cell Growth Differ 1996, 7, 1793–1804. [Google Scholar]
- Providence, K.M.; Kutz, S.M.; Staiano-Coico, L.; Higgins, P.J. PAI-1 gene expression is regionally induced in wounded epithelial cell monolayers and required for injury repair. J. Cell Physiol 2000, 1, 269–280. [Google Scholar]
- Providence, K.M.; Higgins, S.P.; Mullen, A.; Battista, A.; Samarakoon, R.; Higgins, C.E.; Wilkins-Port, C.E.; Higgins, P.J. SERPINE1 (PAI-1) is deposited into keratinocyte migration “trails” and required for optimal monolayer wound repair. Arch. Dermatol. Res 2008, 300, 303–310. [Google Scholar]
- Bandyopadhyay, B.; Fan, J.; Guan, S.; Li, Y.; Chen, M.; Woodley, D.T.; Li, W. A “traffic control” role for TGFbeta3: Orchestrating dermal and epidermal cell motility during wound healing. J. Cell Biol 2006, 172, 1093–1105. [Google Scholar]
- Li, C.M.; Li, W.; Man, X.Y.; Zhou, J.; Chen, J.Q.; Cai, S.Q.; Zheng, M. Pigment epithelium-derived factor plays an inhibitory role in proliferation and migration of HaCaT cells. Mol. Biol. Rep 2011, 38, 2099–2105. [Google Scholar]
- Kim, W.S.; Park, B.S.; Kim, H.K.; Park, J.S.; Kim, K.J.; Choi, J.S.; Choi, J.S.; Chung, S.J.; Kim, D.D.; Sung, J.H. Evidence supporting antioxidant action of adipose-derived stem cells: Protection of human dermal fibroblasts from oxidative stress. J. Dermatol. Sci 2008, 49, 133–142. [Google Scholar]
- Kim, W.S.; Park, B.S.; Sung, J.H.; Yang, J.M.; Park, S.B.; Kwak, S.J.; Park, J.S. Wound healing effect of adipose-derived stem cells: A critical role of secretory factors on human dermal fibroblasts. J. Dermatol. Sci 2007, 48, 15–24. [Google Scholar]
- Li, J.; Chen, J.; Kirsner, R. Pathophysiology of acute wound healing. Clin. Dermatol 2007, 25, 9–18. [Google Scholar]
- Bradford, M.M. Rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem 1976, 72, 248–254. [Google Scholar]
- Oakley, B.R.; Kirsch, D.R.; Morris, N.R. A simplified ultrasensitive silver stain for detecting proteins in polyacrylamide gels. Anal. Biochem 1980, 105, 361–363. [Google Scholar]
- Fernandez, J.; Gharahdaghi, F.; Mische, S.M. Routine identification of proteins from sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-APGE) gels or polyvinyl difluoride membranes using matrix assiated laser desorption/ionization time of flight-mass spectrometry (MALDI-TOF-MS). Electrophoresis 1998, 19, 1036–1045. [Google Scholar]

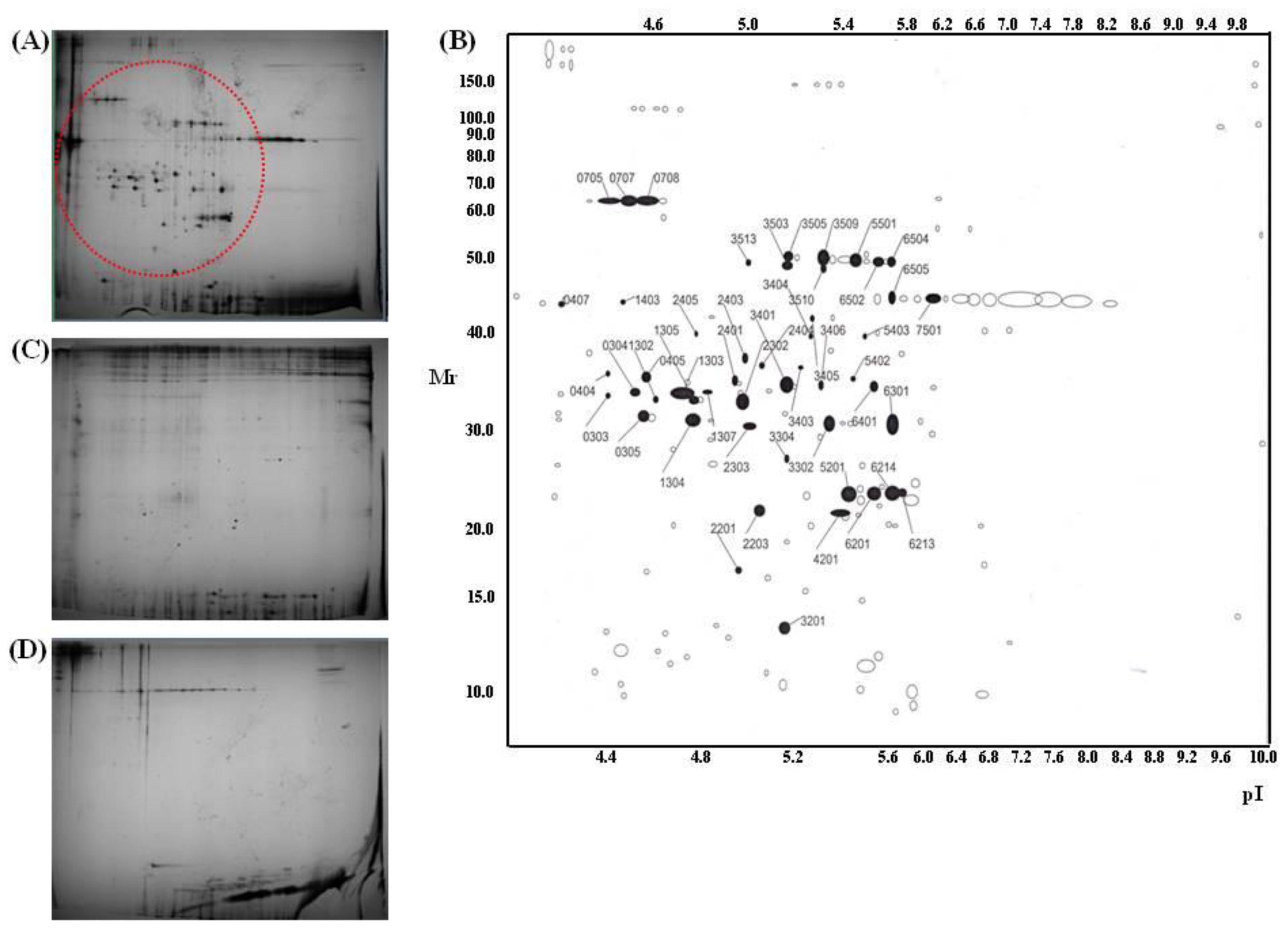
| Trypsin spot | Protein name | Accession # | Protein score | Pep. count | Position | Sequence coverage |
|---|---|---|---|---|---|---|
| 303 | Type VI collagen, alpha-2, isoform 2C2a’ | NP_478055 | 86 | 9 | 96–393 | 12% |
| 304 | Human pro alpha 1(I) collagen | CAA29605 | 138 | 7 | 9–196 | 45% |
| 305 | Type VI collagen, alpha-1 chain | CAA33888 | 210 | 11 | 52–256 | 50% |
| 404 | Type VI collagen, alpha-1 C-terminal domain | CAA33888 | 155 | 8 | 69–211 | 39% |
| 405 | Type VI collagen, alpha-1 C-terminal domain | CAA33888 | 162 | 9 | 52–256 | 41% |
| 1302 | Type VI collagen, alpha 2, isoform | EAX09316 | 103 | 9 | 96–411 | 12% |
| 1303 | Human pro alpha 1(I) collagen | CAA29605 | 142 | 11 | 9–196 | 48% |
| 1304 | Type VI collagen, alpha-1 C-terminal | CAA33888 | 288 | 16 | 28–256 | 67% |
| 1305 | Carboxy-propeptide of alpha 1 (III) procollagen | CAA25879 | 150 | 11 | 1–211 | 45% |
| 1307 | Carboxy-propeptide of alpha 1 (III) procollagen | CAA25879 | 41 | 3 | 46–52 | 18% |
| 2302 | Carboxy-propeptide of alpha 1 (III) procollagen | CAA25879 | 179 | 13 | 1–211 | 43% |
| 2303 | Type VI collagen, alpha-1 C-terminal domain | CAA33888 | 139 | 8 | 52–256 | 30% |
| 2401 | Type I collagen | CAA39142 | 120 | 9 | 165–361 | 25% |
| 2403 | Human pro alpha 1(I) collagen | CAA29605 | 129 | 7 | 9–196 | 45% |
| 2404 | Carboxy-propeptide of alpha 1 (III) procollagen | CAA25879 | 213 | 13 | 43–211 | 47% |
| 3302 | Type I collagen | CAA39142 | 137 | 12 | 165–361 | 29% |
| 3401 | Type I collagen | CAA39142 | 147 | 12 | 165–361 | 29% |
| 3403 | Carboxy-propeptide of alpha 1 (III) procollagen | CAA25879 | 175 | 12 | 43–211 | 44% |
| 3404 | Pro alpha 1(I) collagen | CAA29605 | 85 | 7 | 9–171 | 37% |
| 3405 | Pro alpha 1(I) collagen | CAA29605 | 90 | 8 | 9–171 | 30% |
| 3406 | Type I collagen | CAA39142 | 127 | 9 | 165–361 | 25% |
| 3503 | Type VI collagen, alpha-1 | CAA33889 | 114 | 9 | 91–431 | 23% |
| 3505 | Type VI collagen, alpha-1 | CAA33889 | 148 | 12 | 101–432 | 27% |
| 3509 | Type VI collagen, alpha-1 | CAA33889 | 187 | 13 | 91–432 | 30% |
| 3510 | Type VI collagen, alpha-1 | CAA33889 | 94 | 7 | 120–431 | 20% |
| 3513 | Type VI collagen, alpha-1 | CAA33889 | 97 | 8 | 120–431 | 21% |
| 5403 | Pro alpha 1(I) collagen | CAA29605 | 133 | 10 | 9–196 | 49% |
| 5501 | Type VI collagen, alpha-1 | CAA33889 | 208 | 14 | 91–432 | 31% |
| 6301 | Type I collagen | CAA39142 | 217 | 16 | 165–361 | 41% |
| 6401 | Type I collagen | CAA39142 | 164 | 14 | 165–361 | 29% |
| 6502 | Type VI collagen, alpha-1 | CAA33889 | 134 | 10 | 120–431 | 27% |
| 407 | Human Plasminogen Activator Inhibitor Type-1 | 1A7C_A | 182 | 15 | 1–379 | 42% |
| 705 | Gelatinase A | 1CK7_A | 185 | 18 | 1–631 | 34% |
| 707 | Gelatinase A | 1CK7_A | 249 | 23 | 1–561 | 47% |
| 708 | Gelatinase A | 1CK7_A | 247 | 21 | 1–561 | 40% |
| 2201 | Connective tissue growth factor precursor | NP_001892 | 77 | 5 | 43–152 | 14% |
| 2203 | Interleukin-6 precursor | NP_000591 | 96 | 5 | 59–207 | 22% |
| 2405 | Human procathepsin L | 1CJL_A | 97 | 6 | 18–292 | 23% |
| 3201 | Cadherin 2, isoform | EAX01239 | 64 | 6 | 34–114 | 8% |
| 4201 | Heparan sulfate proteoglycan 2 | EWA94994 | 67 | 11 | 181–3588 | 3% |
| 5201 | Prommp-2TIMP-2 | 1GXD_C | 62 | 4 | 28–194 | 19% |
| 6201 | SPAG9 protein | AAI06049 | 70 | 7 | 1–461 | 21% |
| 6213 | Prommp-2TIMP | 1GXD_C | 68 | 5 | 28–194 | 23% |
| 6214 | Prommp-2TIMP | 1GXD_C | 89 | 7 | 28–194 | 30% |
| 6505 | Chain A of human PEDF | 1IMV_A | 141 | 10 | 34–391 | 24% |
| 7501 | Chain A of human PEDF | 1IMV_A | 161 | 13 | 34–398 | 30% |
| 1403 | N/I * | |||||
| 3304 | N/I | |||||
| 5402 | N/I |
© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Moon, K.M.; Park, Y.-H.; Lee, J.S.; Chae, Y.-B.; Kim, M.-M.; Kim, D.-S.; Kim, B.-W.; Nam, S.-W.; Lee, J.-H. The Effect of Secretory Factors of Adipose-Derived Stem Cells on Human Keratinocytes. Int. J. Mol. Sci. 2012, 13, 1239-1257. https://doi.org/10.3390/ijms13011239
Moon KM, Park Y-H, Lee JS, Chae Y-B, Kim M-M, Kim D-S, Kim B-W, Nam S-W, Lee J-H. The Effect of Secretory Factors of Adipose-Derived Stem Cells on Human Keratinocytes. International Journal of Molecular Sciences. 2012; 13(1):1239-1257. https://doi.org/10.3390/ijms13011239
Chicago/Turabian StyleMoon, Kyoung Mi, Ye-Hyoung Park, Jae Seol Lee, Yong-Byung Chae, Moon-Moo Kim, Dong-Soo Kim, Byung-Woo Kim, Soo-Wan Nam, and Jong-Hwan Lee. 2012. "The Effect of Secretory Factors of Adipose-Derived Stem Cells on Human Keratinocytes" International Journal of Molecular Sciences 13, no. 1: 1239-1257. https://doi.org/10.3390/ijms13011239
APA StyleMoon, K. M., Park, Y.-H., Lee, J. S., Chae, Y.-B., Kim, M.-M., Kim, D.-S., Kim, B.-W., Nam, S.-W., & Lee, J.-H. (2012). The Effect of Secretory Factors of Adipose-Derived Stem Cells on Human Keratinocytes. International Journal of Molecular Sciences, 13(1), 1239-1257. https://doi.org/10.3390/ijms13011239




