FurinDB: A Database of 20-Residue Furin Cleavage Site Motifs, Substrates and Their Associated Drugs
Abstract
:1. Introduction
2. Results and Discussion
2.1. Database Content
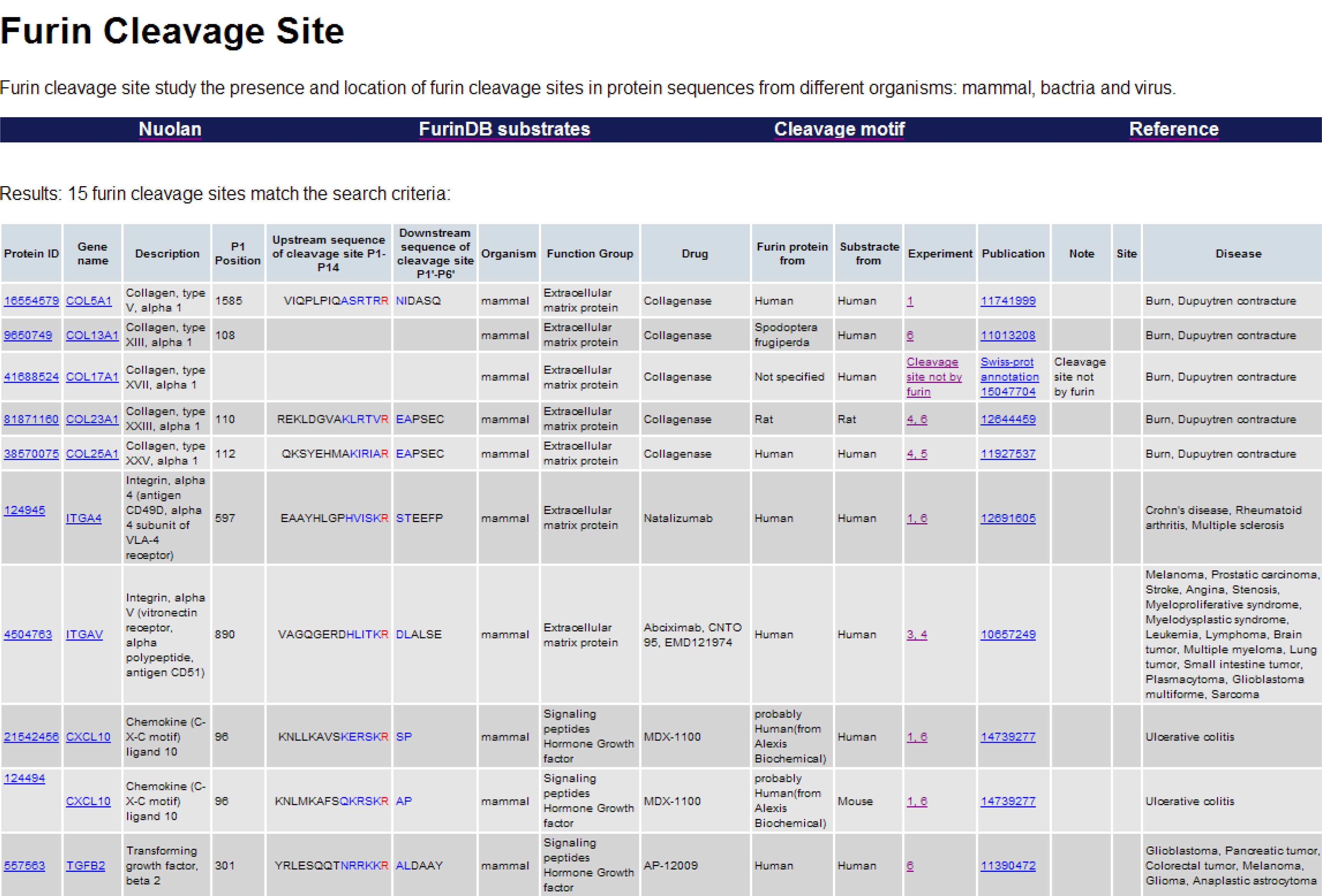
- Furin substrate sequence information: Name and description of the gene encoding the substrate protein and the substrate protein ID. External links to the NCBI protein sequence database and the NCBI Entrez gene database are also provided.
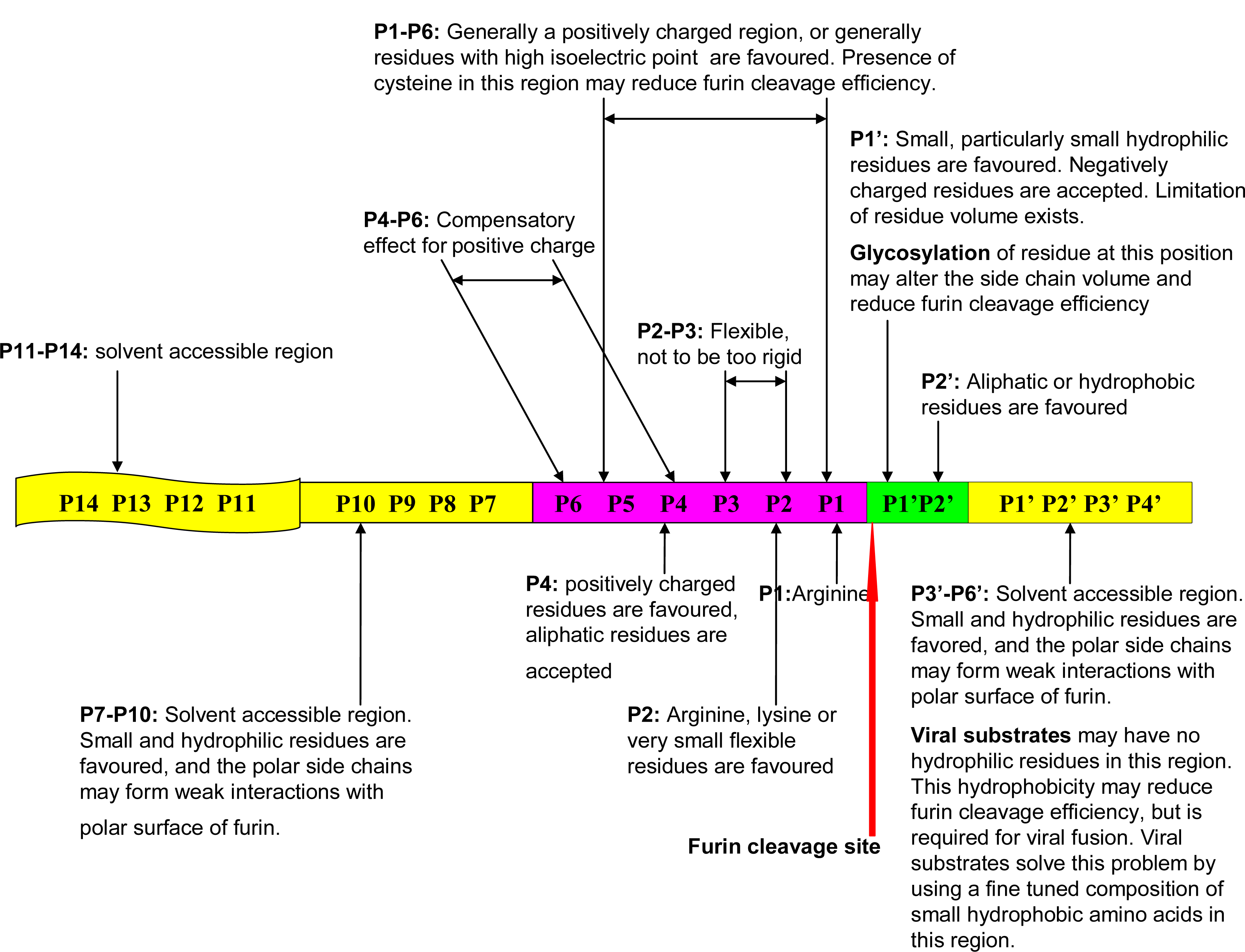
- Furin cleavage site sequence information and location: The furin cleavage site P14–P6′ is represented as a 20 amino acids motif that contains one core region (eight amino acids, positions P6–P2′) packed inside the furin binding pocket; and two polar regions (eight amino acids, positions P7–P14; and four amino acids, positions P3′–P6′) located outside of the furin binding pocket [4]. The position P1, immediately prior to the furin cleavage site is recorded. The presence of multiple cleavage sites on the same protein substrate is indicated.
- Taxonomy information of substrate: Each furin substrate is assigned to one of three taxonomy groups (mammals, bacteria or viruses).
- Functional information about the furin substrates: Each furin substrate is assigned to one of seven predefined biological functions: (1) extracellular matrix protein; (2) signal peptide, hormone or growth factor; (3) serum protein; (4) transmembrane receptor; (5) viral protein; (6) bacterial protein or (7) other [1,2].
- Experimental methods and conditions: Methods used to measure furin cleavage and the final cleavage products, the origin of the furin enzyme and the origin of the furin substrate used in the experiment and the literature record of the original experiments. These detailed experimental methods and conditions are grouped into 10 different types [5]. External links to the PUBMED literature database are also provided.
- Associated drugs: Information is included for each furin substrate targeted by a drug that has either entered phase I trials or has already received FDA approval.
2.2. Database Access and Web Interface
- Query by seven functional categories, three species and if the furin substrate is a known drug target.
- Query by standard Entrez gene name.
- Query by the eight amino acids motif in the core region P6–P2′ of the furin cleavage site, which is located inside the furin binding pocket. Either one or multiple amino acids can be input at any of the eight positions. The symbol X is interpreted to mean any amino acid.
3. Future Development
4. Conclusions
Acknowledgments
References
- Molloy, SS; Anderson, ED; Jean, F; Thomas, G. Bi-cycling the furin pathway: from TGN localization to pathogen activation and embryogenesis. Trends Cell Biol 1999, 9, 28–35. [Google Scholar]
- Thomas, G. Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat. Rev. Mol. Cell Biol 2002, 3, 753–766. [Google Scholar]
- Hajdin, K; D'Alessandro, V; Niggli, FK; Schafer, BW; Bernasconi, M. Furin targeted drug delivery for treatment of rhabdomyosarcoma in a mouse model. PLoS One 2010, 5, e10445. [Google Scholar]
- Sun, T. A 20 Residues Motif Delineates the Furin Cleavage Site and its Physical Properties May Influence Viral Fusion. Biochem. Insights 2009, 2, 9–20. [Google Scholar]
- Furin Cleavage Site. Available online: http://www.nuolan.net/substrates.html (accessed on 7 February 2011).
- Nakayama, K. Furin: a mammalian subtilisin/Kex2p-like endoprotease involved in processing of a wide variety of precursor proteins. Biochem. J 1997, 327, 625–635. [Google Scholar]
- Seidah, NG; Chretien, M. Proprotein and prohormone convertases: a family of subtilases generating diverse bioactive polypeptides. Brain Res 1999, 848, 45–62. [Google Scholar]
- Sun, T; Wu, J. Comparative study of the binding pockets of mammalian proprotein convertases and its implications for the design of specific small molecule inhibitors. Int. J. Biol. Sci 2010, 6, 89–95. [Google Scholar]
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Tian, S.; Huang, Q.; Fang, Y.; Wu, J. FurinDB: A Database of 20-Residue Furin Cleavage Site Motifs, Substrates and Their Associated Drugs. Int. J. Mol. Sci. 2011, 12, 1060-1065. https://doi.org/10.3390/ijms12021060
Tian S, Huang Q, Fang Y, Wu J. FurinDB: A Database of 20-Residue Furin Cleavage Site Motifs, Substrates and Their Associated Drugs. International Journal of Molecular Sciences. 2011; 12(2):1060-1065. https://doi.org/10.3390/ijms12021060
Chicago/Turabian StyleTian, Sun, Qingsheng Huang, Ying Fang, and Jianhua Wu. 2011. "FurinDB: A Database of 20-Residue Furin Cleavage Site Motifs, Substrates and Their Associated Drugs" International Journal of Molecular Sciences 12, no. 2: 1060-1065. https://doi.org/10.3390/ijms12021060



