Molecular Interactions and Protein-Induced DNA Hairpin in the Transcriptional Control of Bacteriophage Ø29 DNA
Abstract
:1. Introduction
2. Molecular Requirements in the Transcriptional Switch from Early to Late Gene Expression During Bacteriophage Ø29 Infection
3. Protein p4-DNA Complex: Direct and Indirect-Readout Mechanisms Involved in the Recognition of Target Sequences
4. Zipper Model for p4 Specific Sequence Recognition and DNA Binding
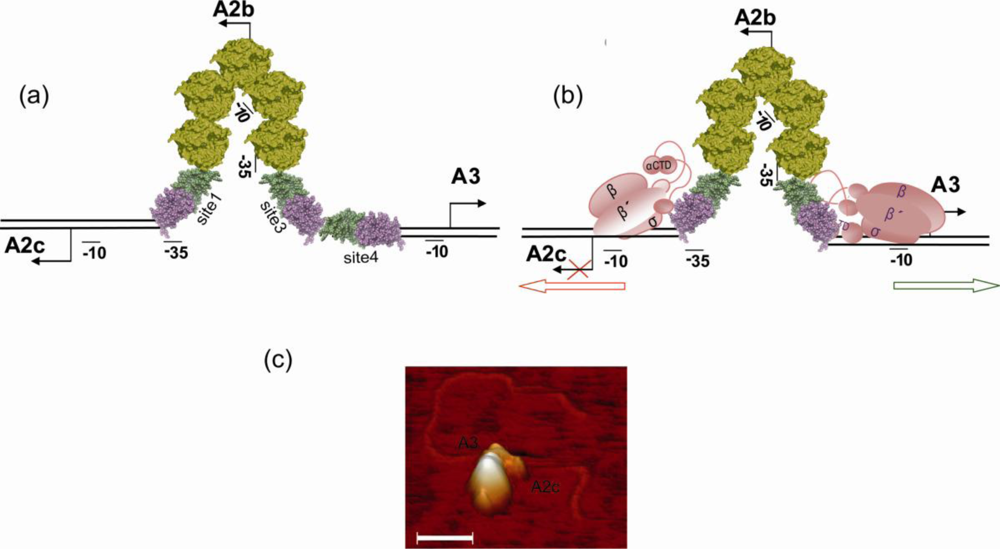
5. Regulation of the Switch from Early to Late Gene Expression
6. Conclusions
Acknowledgments
References
- Koo, BM; Rhodius, VA; Nonaka, G; de Haseth, PL; Gross, CA. Reduced capacity of alternative sigmas to melt promoters ensures stringent promoter recognition. Genes Dev 2009, 23, 2426–2436. [Google Scholar]
- Hughes, KT; Mathee, K. The anti-sigma factors. Annu. Rev. Microbiol 1998, 52, 231–286. [Google Scholar]
- Yuan, AH; Nickels, BE; Hochschild, A. The bacteriophage T4 AsiA protein contacts the {beta}-flap domain of RNA polymerase. Proc. Natl. Acad. Sci. USA 2009, 106, 6597–6602. [Google Scholar]
- Gilmore, JM; Urbauer, RJ; Minakhin, L; Akoyev, V; Zolkiewski, M; Severinov, KV; Urbauer, JL. Determinants of Affinity and Activity of the Anti-Sigma Factor AsiA. Biochemistry 2010. [Google Scholar] [CrossRef]
- Campbell, EA; Westblade, LF; Darst, SA. Regulation of bacterial RNA polymerase sigma factor activity: a structural perspective. Curr. Opin. Microbiol 2008, 11, 121–127. [Google Scholar]
- Barnard, A; Wolfe, A; Busby, S. Regulation at complex bacterial promoters: how bacteria use different promoter organizations to produce different regulatory outcomes. Curr. Opin. Microbiol 2004, 7, 102–108. [Google Scholar]
- Dorman, CJ; Deighan, P. Regulation of gene expression by histone-like proteins in bacteria. Curr. Opin. Genet. Dev 2003, 13, 179–184. [Google Scholar]
- Rimsky, S. Structure of the histone-like protein H-NS and its role in regulation and genome superstructure. Curr. Opin. Microbiol 2004, 7, 109–114. [Google Scholar]
- Fang, FC; Rimsky, S. New insights into transcriptional regulation by H-NS. Curr. Opin. Microbiol 2008, 11, 113–120. [Google Scholar]
- Heffernan, L; Wilcox, G. Effect of araC gene product on catabolite repression in the L-arabinose regulon. J. Bacteriol 1976, 126, 1132–1135. [Google Scholar]
- Adhya, S. Multipartite genetic control elements: communication by DNA loop. Annu. Rev. Genet 1989, 23, 227–250. [Google Scholar]
- Choy, HE; Park, SW; Aki, T; Parrack, P; Fujita, N; Ishihama, A; Adhya, S. Repression and activation of transcription by Gal and Lac repressors: involvement of α subunit of RNA polymerase. EMBO J 1995, 14, 4523–4529. [Google Scholar]
- Hochschild, A; Dove, SL. Protein-protein contacts that activate and repress prokaryotic transcription. Cell 1998, 92, 597–600. [Google Scholar]
- Schleif, R. Regulation of the L-arabinose operon of Escherichia coli. Trends Genet 2000, 16, 559–565. [Google Scholar]
- Lloyd, GP; Landini, P; Busby, S. Activator and repression of transcription initiation in bacteria. Essay Biochem 2001, 37, 17–31. [Google Scholar]
- Scaffidi, P; Bianchi, ME. Spatially precise DNA bending is an essential activity of the Sox2 transcription factor. J. Biol. Chem 2001, 276, 47296–47302. [Google Scholar]
- Lim, FL; Hayes, A; West, AG; Pic-Taylor, A; Darieva, Z; Morgan, BA; Oliver, SG; Sharrocks, AD. Mcm1p-induced DNA bending regulates the formation of ternary transcription factor complex. Mol. Cell. Biol 2003, 23, 450–461. [Google Scholar]
- Yang, J; Hwang, JS; Camakaris, H; Irawaty, W; Ishihama, A; Pittard, J. Mode of action of the TyrR protein: repression and activation of the tyrP promoter of Escherichia coli. Mol. Microbiol 2004, 52, 243–256. [Google Scholar]
- Browning, DF; Busby, SJW. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol 2004, 2, 57–65. [Google Scholar]
- van Hijum, SA; Medema, MH; Kuipers, OP. Mechanisms and evolution of control logic in prokaryotic transcriptional regulation. Microbiol. Mol. Biol. Rev 2009, 73, 481–509. [Google Scholar]
- Hochschild, A; Lewis, M. The bacteriophage lambda CI protein finds an asymmetric solution. Curr. Opin. Struct. Biol 2009, 19, 79–86. [Google Scholar]
- Pittard, J; Camakaris, H; Yang, J. The TyrR regulon. Mol. Microbiol 2005, 55, 16–26. [Google Scholar]
- Hochschild, A; Ptashne, M. Cooperative binding of lambda repressors to sites separated by integral turns of the DNA helix. Cell 1986, 44, 681–687. [Google Scholar]
- Schreiber, V; Steegborn, C; Clausen, T; Boos, W; Richet, E. A new mechanism for the control of a prokaryotic transcriptional regulator: antagonistic binding of positive and negative effectors. Mol. Microbiol 2000, 35, 765–776. [Google Scholar]
- Joly, N; Bohm, A; Richet, E. MalK, the Atp-binding casset component of the Escherichia coli Maltodextrin transporter, inhibits the transcriptional activator MalT and antagonizing inducer binding. J. Biol. Chem 2004, 279, 33123–33130. [Google Scholar]
- Squire, DJ; Xu, M; Cole, JA; Busby, SJ; Browning, DF. Competition between NarL-dependent activation and Fis-dependent repression controls expression from the Escherichia coli yeaR and ogt promoters. Biochem. J 2009, 420, 249–257. [Google Scholar]
- Grosschedl, R. Higher-order nucleoprotein complexes in transcription: analogies with site-specific recombination. Curr. Opin. Cell Biol 1995, 7, 362–370. [Google Scholar]
- Pérez-Martin, J; de Lorenzo, V. Clues and consequences of DNA bending in transcription. Annu. Rev. Microbiol 1997, 51, 593–628. [Google Scholar]
- Bourgerie, SJ; Michan, CM; Thomas, MS; Busby, SJ; Hyde, EI. DNA binding and DNA bending by the MelR transcription activator protein from Escherichia coli. Nucleic Acids Res 1997, 25, 1685–1693. [Google Scholar]
- Kahn, JD; Crothers, DM. Measurement of the DNA bend angle induced by the catabolite activator protein using Monte Carlo simulation of cyclization kinetics. J. Mol. Biol 1998, 276, 287–309. [Google Scholar]
- Ross, ED; Keating, AM; Maher, LJ. DNA constraints on transcription activation in vitro. J. Mol. Biol 2000, 297, 321–334. [Google Scholar]
- Lewis, DE; Lee, HJ; Liu, M; Adhya, S. Effect of varying the supercoiling of DNA on transcription and its regulation. Biochemstry 2003, 42, 10718–10725. [Google Scholar]
- Kim, SI; Jourlin-Caselli, C; Wellington, SR; Sonenshein, AL. Mechanism of repression by Bacillus subtilis CcpC, a LysR family regulator. J. Mol. Biol 2003, 334, 609–624. [Google Scholar]
- Loskutoff, DJ; Pène, JJ. Gene expression during the development of Bacillus subtilis bacteriophage Ø29. I. Analysis of viral-specific transcription by deoxyribonucleic acid-ribonucleic acid competition hybridization. J. Virol 1973, 11, 78–86. [Google Scholar]
- Sogo, JM; Inciarte, MR; Corral, J; Viñuela, E; Salas, M. RNA polymerase binding sites and transcription map of the DNA of Bacillus subtilis phage Ø29. J. Mol. Biol 1979, 127, 411–436. [Google Scholar]
- Chang, BY; Shyu, YT; Doi, RH. The interaction between Bacillus subtilis σA (sigma A) factor and RNA polymerase with promoters. Biochimie 1992, 74, 601–612. [Google Scholar]
- Helmann, JD. Compilation and analysis of Bacillus subtilis σA-dependent promoter sequences: evidence for extended contact between RNA polymerase and upstream promoter DNA. Nucleic Acids Res 1995, 23, 2351–2360. [Google Scholar]
- Voskuil, MI; Voepel, K; Chambliss, GH. The −16 region, a vital sequence for the utilization of a promoter in Bacillus subtilis and Escherichia coli. Mol. Microbiol 1995, 17, 271–279. [Google Scholar]
- Minakhin, L; Bhagat, S; Brunning, A; Campbell, EA; Darst, SA; Ebright, RH; Severinov, K. Bacterial RNA polymerase subunit omega and eukaryotic RNA polymerase subunit RPB6 are sequence, structural, and functional homologs and promote RNA polymerase assembly. Proc. Natl. Acad. Sci. USA 2001, 98, 892–897. [Google Scholar]
- Ebright, RH. RNA polymerase: structural similarities between bacterial RNA polymerase and eukaryotic RNA polymerase II. J. Mol. Biol 2002, 304, 687–698. [Google Scholar]
- Gruber, TM; Gross, CA. Multiple σ subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol 2003, 57, 441–466. [Google Scholar]
- Mathew, R; Chatterji, D. The evolving story of the omega subunit of bacterial RNA polymerase. Trends Microbiol 2006, 14, 450–455. [Google Scholar]
- Campbell, EA; Westblade, LF; Darst, SA. Regulation of bacterial RNA polymerase sigma factor activity: a structural perspective. Curr. Opin. Microbiol 2008, 11, 121–127. [Google Scholar]
- Rojo, F; Barthelemy, I; Nuez, B; Serrano, M; Salas, M. Transcription regulation in Bacillus subtilis phage Ø29. Res. Microbiol 1991, 142, 771–777. [Google Scholar]
- Guo, P; Erickson, S; Anderson, D. A small viral RNA is required for in vitro packaging of bacteriophage Ø29 DNA. Science 1987, 236, 690–694. [Google Scholar]
- Barthelemy, I; Salas, M; Mellado, RP. In vivo transcription of bacteriophage Ø29 DNA: transcription termination. J. Virol 1987, 61, 1751–1755. [Google Scholar]
- Camacho, A; Salas, M. DNA bending and looping in the transcriptional control of bacteriophage Ø29. FEMS Microbiol. Rev 2010, 34, 828–841. [Google Scholar]
- Monsalve, M; Mencía, M; Rojo, F; Salas, M. Transcription regulation in Bacillus subtilis phage Ø29: expression of the viral promoters throughout the infection cycle. Virology 1995, 207, 23–31. [Google Scholar]
- Camacho, A; Salas, M. Pleiotropic effect of protein p6 on the viral cycle of bacteriophage Ø29. J. Bacteriol 2000, 182, 6927–6932. [Google Scholar]
- Serrano, M; Salas, M; Hermoso, JM. A novel nucleoprotein complex at a replication origin. Science 1990, 248, 1012–1016. [Google Scholar]
- González-Huici, V; Alcorlo, M; Salas, M; Hermoso, JM. Bacteriophage Ø29 protein p6: an architectural protein involved in genome organization, replication and control of transcription. J. Mol. Recognit 2004, 17, 390–396. [Google Scholar]
- Barthelemy, I; Mellado, RP; Salas, M. In vitro transcription of bacteriophage Ø29 DNA: inhibition of early promoters by the viral replication protein p6. J. Virol 1989, 63, 460–462. [Google Scholar]
- Camacho, A; Salas, M. Repression of bacteriophage Ø29 early promoter C2 by viral protein p6 is due to impairment of closed complex. J. Biol. Chem 2001, 276, 28927–28932. [Google Scholar]
- Elías-Arnanz, M; Salas, M. Functional interactions between a phage histone-like protein and a transcriptional factor in regulation of Ø29 early-late transcriptional switch. Genes Dev 1999, 13, 2502–2513. [Google Scholar]
- Camacho, A; Salas, M. Mechanism for the switch of Ø29 DNA early to late transcription by regulatory protein p4 and histone-like protein p6. EMBO J 2001, 20, 6060–6070. [Google Scholar]
- Drew, HR; Travers, AA. DNA bending and its relation to nucleosome positioning. J. Mol. Biol 1985, 186, 773–790. [Google Scholar]
- Kalodimos, CG; Biris, N; Bonvin, AM; Levandoski, MM; Guennuegues, M; Boelens, R; Kaptein, R. Structure and flexibility adaptation in nonspecific and specific protein-DNA complexes. Science 2004, 305, 386–389. [Google Scholar]
- Napoli, AA; Lawson, CL; Ebright, RH; Berman, HM. Indirect readout of DNA sequence at the primary-kink site in the CAP-DNA complex: recognition of pyrimidine-purine and purine-purine steps. J. Mol. Biol 2006, 357, 173–183. [Google Scholar]
- Kouldelka, GB; Mauro, SA; Ciubotaru, M. Indirect readout of DNA sequence by proteins: The roles of DNA sequence dependent intrinsic and extrinsic forces. Prog. Nucleic Acid Res. Mol. Biol 2007, 81, 143–177. [Google Scholar]
- Hogan, ME; Austin, RH. Importance of DNA stiffness in protein-DNA binding specificity. Nature 1987, 329, 263–266. [Google Scholar]
- von Hippel, PH. Protein-DNA recognition: new perspectives and underlying themes. Science 1994, 263, 769–770. [Google Scholar]
- Watkins, D; Hsiao, C; Woods, KK; Koudelka, GB; Williams, LD. P22 c2 repressor-operator complex: mechanisms of direct and indirect readout. Biochemistry 2008, 47, 2325–2338. [Google Scholar]
- Rohs, R; West, SM; Sosinsky, A; Liu, P; Mann, RS; Honig, B. The role of DNA shape in protein-DNA recognition. Nature 2009, 461, 1248–1253. [Google Scholar]
- Barthelemy, I; Salas, M. Characterization of a new prokaryotic transcriptional activator and its DNA recognition site. J. Mol. Biol 1989, 208, 225–232. [Google Scholar]
- Nuez, B; Rojo, F; Salas, M. Phage Ø29 regulatory protein p4 stabilizes the binding of the RNA polymerase to the late promoter in a process involving direct protein-protein contact. Proc. Natl. Acad. Sci. USA 1992, 89, 11401–11405. [Google Scholar]
- Nuez, B; Rojo, F; Salas, M. Requirement for an A-tract structure at the binding site of phage Ø29 transcriptional activator. J. Mol. Biol 1994, 237, 175–181. [Google Scholar]
- Pérez-Lago, L; Salas, M; Camacho, A. A precise DNA bend angle is essential for the function of the phage Ø29 transcriptional regulator. Nucleic Acids Res 2005, 33, 126–134. [Google Scholar]
- Badia, D; Camacho, A; Pérez-Lago, L; Escandón, C; Salas, M; Coll, M. The structure of phage Ø29 transcription regulator p4-DNA complex reveals an N-hook motif for DNA. Mol. Cell 2006, 22, 73–81. [Google Scholar]
- Holm, L; Sander, C. Decision support system for the evolutionary classification of protein structures. Proc. Int. Conf. Intell. Syst. Mol. Biol 1997, 5, 140–146. [Google Scholar]
- Mendieta, J; Pérez-Lago, L; Salas, M; Camacho, A. DNA sequence-specific recognition by a transcriptional regulator requires indirect readout of A-tracts. Nucleic Acids Res 2007, 35, 3252–3261. [Google Scholar]
- Mendieta, J; Pérez-Lago, L; Salas, M; Camacho, A. Unpublished work,. 2010.
- Davey, CA; Sargent, DF; Luge, K; Maeder, AW; Richmond, TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J. Mol. Biol 2002, 319, 1097–1113. [Google Scholar]
- Garcia-Pérez, M; Pinto, M; Subirana, JA. Nonsequence-specific arginine interactions in the nucleosome core particle. Biopolymers 2003, 69, 432–439. [Google Scholar]
- Aggarwal, AK; Rodgers, DW; Drottar, M; Ptashne, M; Harrison, SC. Recognition of a DNA operator by the repressor of phage 434: a view at high resolution. Science 1988, 242, 899–907. [Google Scholar]
- Wang, D; Ulyanov, NB; Zhurkin, VB. Sequence-dependent Kink-and-Slide deformations of nucleosomal DNA facilitated by histone arginines bound in the minor groove. J. Biomol. Struct. Dyn 2010, 27, 5843–5980. [Google Scholar]
- Camacho, A; Salas, M. Molecular interplay between RNA polymerase and two transcriptional regulators in promoter switch. J. Mol. Biol 2004, 336, 357–368. [Google Scholar]
- Nuez, B; Rojo, F; Salas, M. Phage Ø29 regulatory protein p4 stabilizes the binding of the RNA polymerase to the late promoter in a process involving direct protein-protein contact. Proc. Natl. Acad. Sci. USA 1992, 89, 11401–11405. [Google Scholar]
- Gutiérrez del Arroyo, PG; Vélez, M; Piétrement, O; Salas, M; Carrascosa, JL; Camacho, A. A nucleoprotein-hairpin in transcription regulation. J. Struct. Biol 2009, 168, 444–451. [Google Scholar]
- Mencía, M; Monsalve, M; Rojo, F; Salas, M. Transcription activation by phage Ø29 protein p4 is mediated by interaction with the α subunit of Bacillus subtilis RNA polymerase. Proc. Natl. Acad. Sci. USA 1996, 93, 6616–6620. [Google Scholar]
- Mencía, M; Monsalve, M; Salas, M; Rojo, F. Transcriptional activator of phage Ø29 late promoter: mapping of residues involved in interaction with RNA polymerase and in DNA bending. Mol. Microbiol 1996, 20, 273–282. [Google Scholar]



© 2010 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Camacho, A.; Salas, M. Molecular Interactions and Protein-Induced DNA Hairpin in the Transcriptional Control of Bacteriophage Ø29 DNA. Int. J. Mol. Sci. 2010, 11, 5129-5142. https://doi.org/10.3390/ijms11125129
Camacho A, Salas M. Molecular Interactions and Protein-Induced DNA Hairpin in the Transcriptional Control of Bacteriophage Ø29 DNA. International Journal of Molecular Sciences. 2010; 11(12):5129-5142. https://doi.org/10.3390/ijms11125129
Chicago/Turabian StyleCamacho, Ana, and Margarita Salas. 2010. "Molecular Interactions and Protein-Induced DNA Hairpin in the Transcriptional Control of Bacteriophage Ø29 DNA" International Journal of Molecular Sciences 11, no. 12: 5129-5142. https://doi.org/10.3390/ijms11125129




