Preclinical Evaluation of a New Series of Albumin-Binding 177Lu-Labeled PSMA-Based Low-Molecular-Weight Radiotherapeutics
Abstract
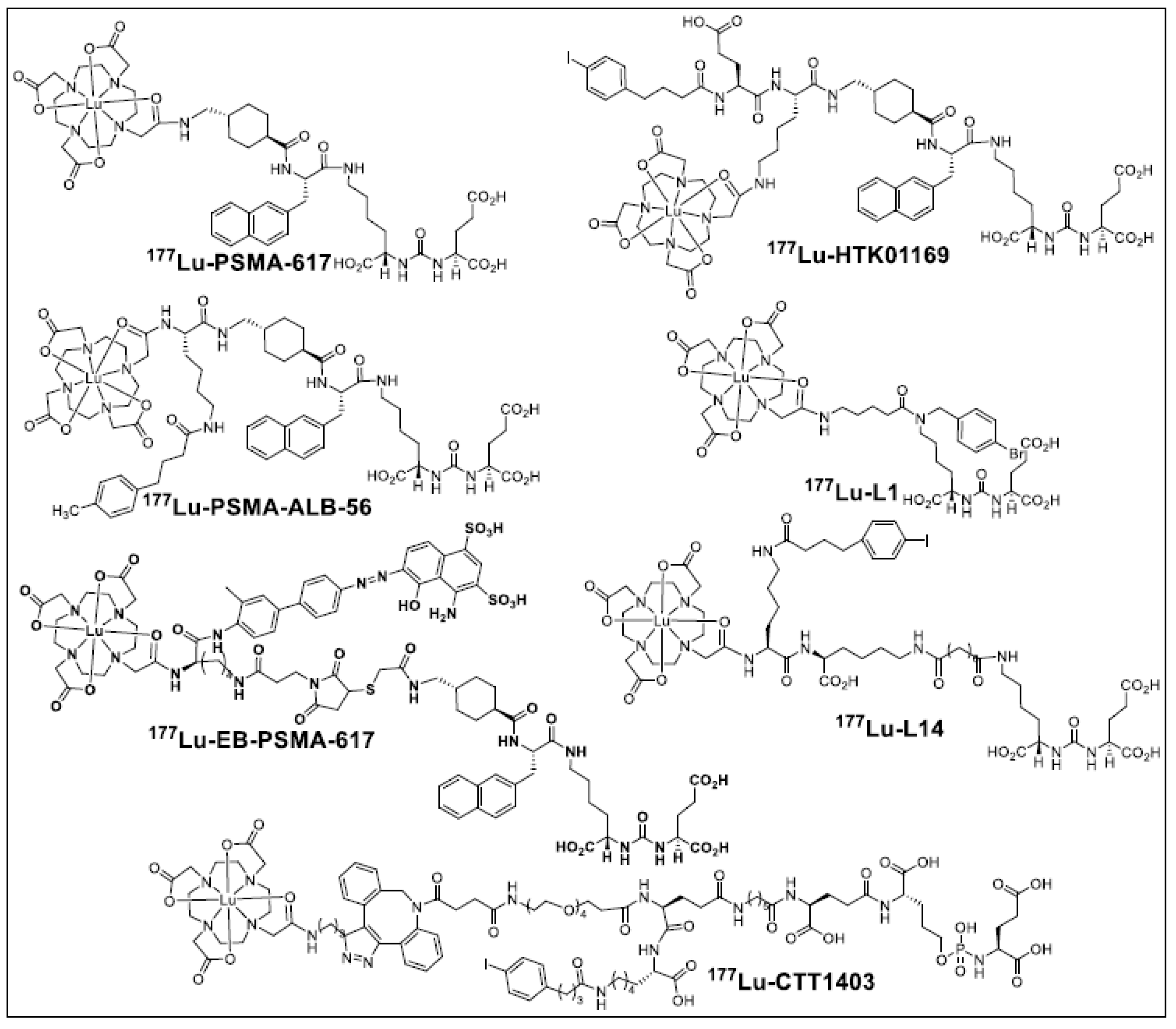
:1. Introduction
2. Results
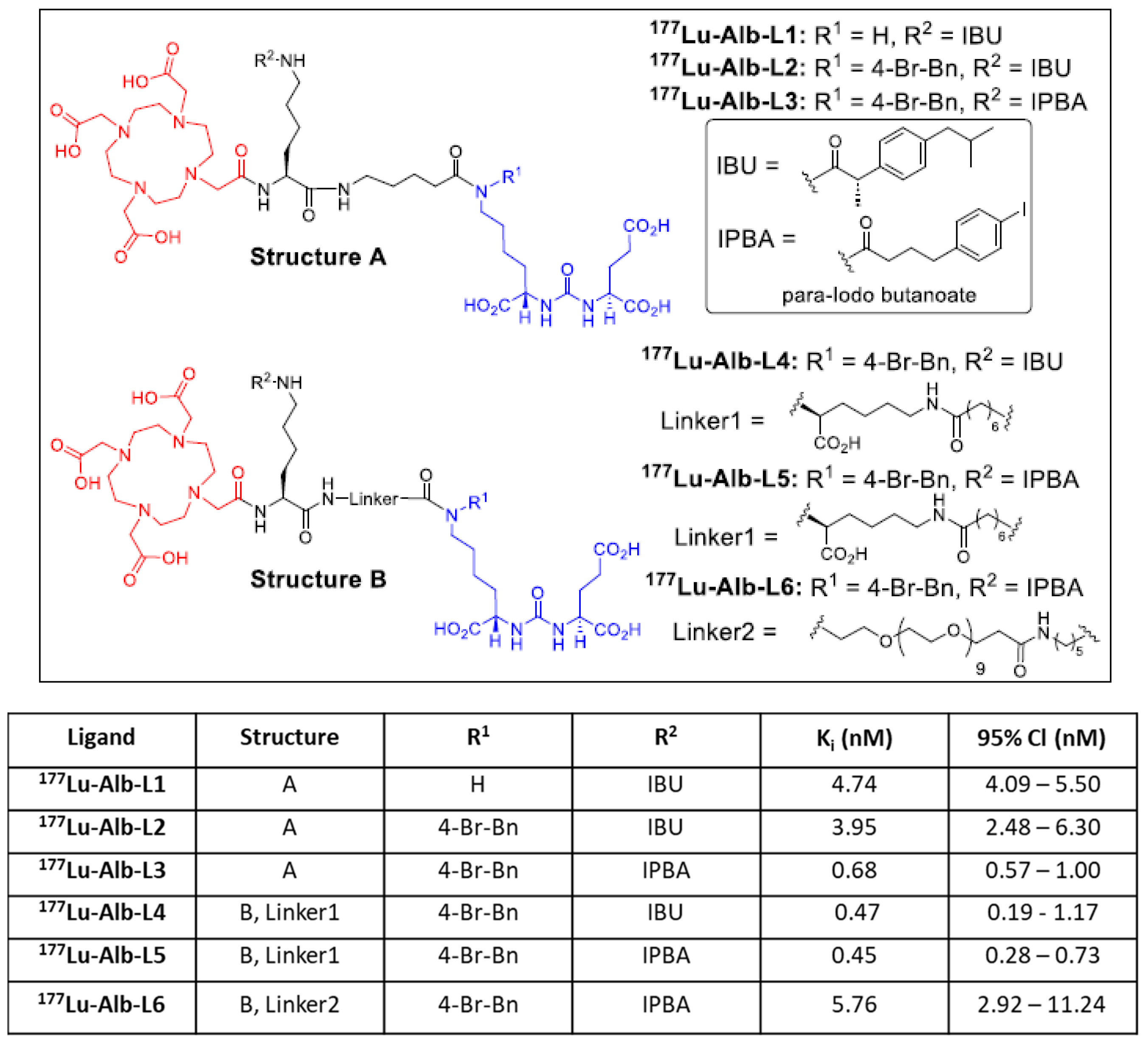
2.1. Synthesis and Binding Affinities of Ligands Alb-L1 to Alb-L6
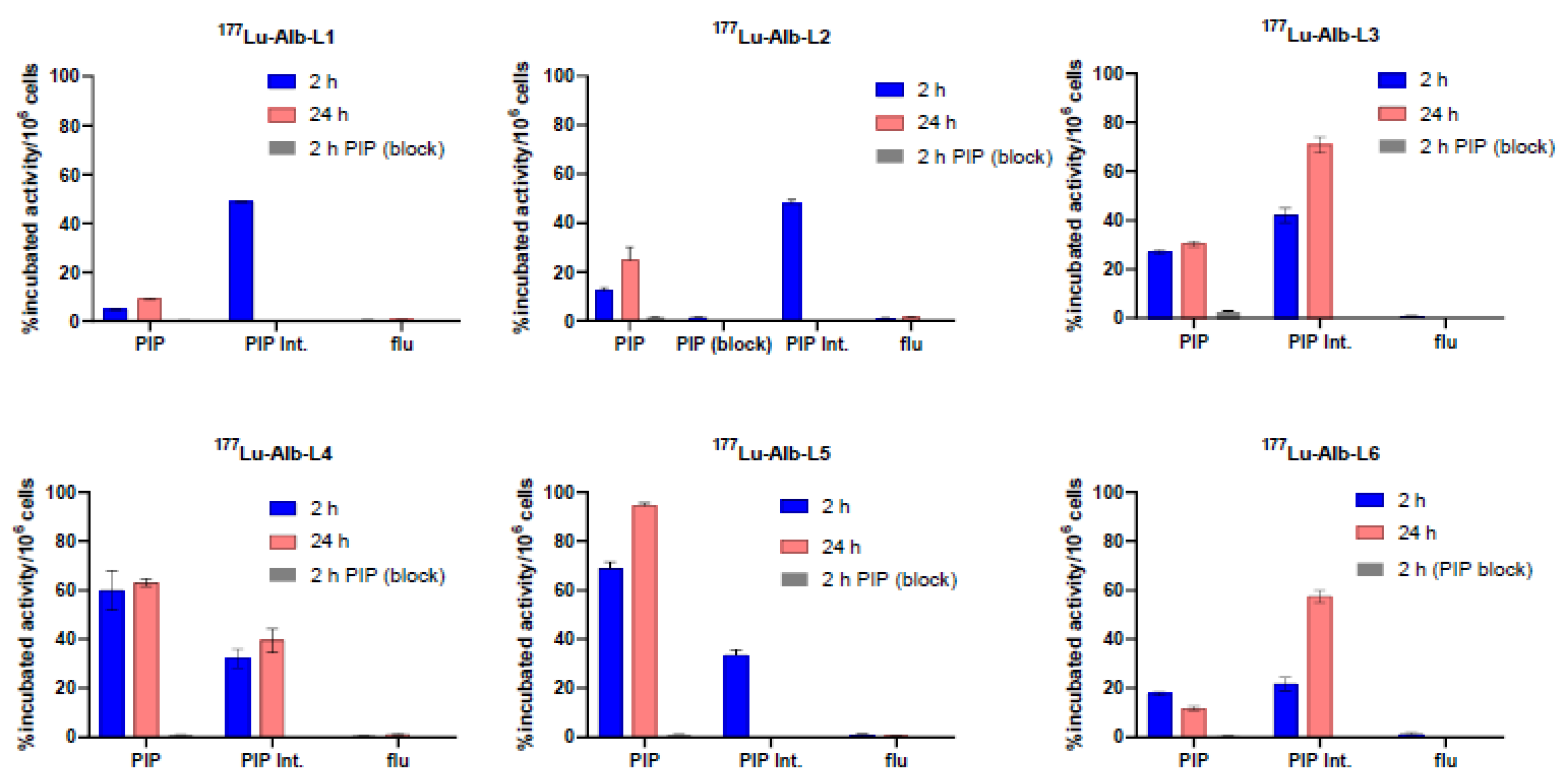
2.2. In Vitro Characterization
Binding Affinity, Cell Uptake, and Internalization
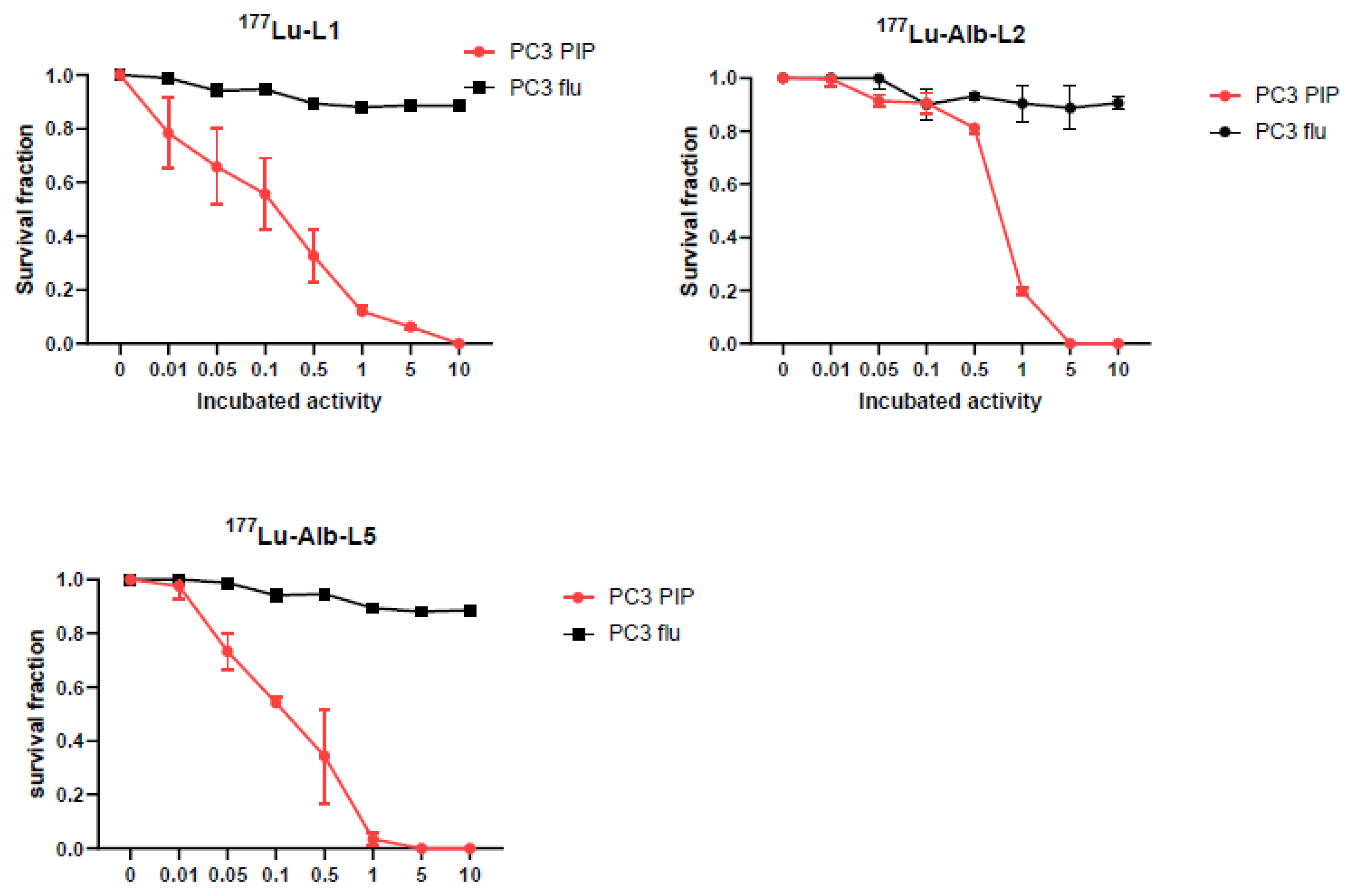
2.3. Clonogenic Survival Assay
2.4. Biodistribution
3. Discussion
4. Materials and Methods
4.1. Radiochemistry
4.2. Measurement of Partition Coefficients
4.3. In Vitro Assays
4.3.1. Competitive Inhibition Assays
4.3.2. Protein-Binding Assay
4.3.3. Cell Culture
4.3.4. Cell Uptake and Internalization Study
4.3.5. Clonogenic Survival Assay
4.4. In Vivo Experiments
Biodistribution
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Rebello, R.J.; Oing, C.; Knudsen, K.E.; Loeb, S.; Johnson, D.C.; Reiter, R.E.; Gillessen, S.; Van der Kwast, T.; Bristow, R.G. Prostate cancer. Nat. Rev. Dis. Primers 2021, 7, 9. [Google Scholar] [CrossRef] [PubMed]
- Sandhu, S.; Moore, C.M.; Chiong, E.; Beltran, H.; Bristow, R.G.; Williams, S.G. Prostate cancer. Lancet 2021, 398, 1075–1090. [Google Scholar] [CrossRef] [PubMed]
- Silver, D.A.; Pellicer, I.; Fair, W.R.; Heston, W.D.; Cordon-Cardo, C. Prostate-specific membrane antigen expression in normal and malignant human tissues. Clin. Cancer Res. 1997, 3, 81–85. [Google Scholar] [PubMed]
- Wright, G.L.; Haley, C.; Beckett, M.L.; Schellhammer, P.F. Expression of prostate-specific membrane antigen in normal, benign, and malignant prostate tissues. Urol. Oncol. Semin. Orig. Investig. 1995, 1, 18–28. [Google Scholar] [CrossRef] [PubMed]
- Sartor, O.; de Bono, J.; Chi, K.N.; Fizazi, K.; Herrmann, K.; Rahbar, K.; Tagawa, S.T.; Nordquist, L.T.; Vaishampayan, N.; El-Haddad, G.; et al. Lutetium-177–PSMA-617 for Metastatic Castration-Resistant Prostate Cancer. N. Eng. J. Med. 2021, 385, 1091–1103. [Google Scholar] [CrossRef]
- Hofman, M.S.; Emmett, L.; Sandhu, S.; Iravani, A.; Joshua, A.M.; Goh, J.C.; Pattison, D.A.; Tan, T.H.; Kirkwood, I.D.; Ng, S.; et al. 177Lu-PSMA-617 versus cabazitaxel in patients with metastatic castration-resistant prostate cancer (TheraP): A randomised, open-label, phase 2 trial. Lancet 2021, 397, 797–804. [Google Scholar] [CrossRef]
- Sandhu, S.; Guo, C.; Hofman, M.S. Radionuclide Therapy in Prostate Cancer: From Standalone to Combination PSMA Theranostics. J. Nucl. Med. 2021, 62, 1660–1668. [Google Scholar] [CrossRef]
- Banerjee, S.R.; Pullambhatla, M.; Shallal, H.; Lisok, A.; Mease, R.C.; Pomper, M.G. A Modular Strategy to Prepare Multivalent Inhibitors of Prostate-Specific Membrane Antigen (PSMA). Oncotarget 2011, 2, 1244. [Google Scholar] [CrossRef]
- Kopka, K.; Benešová, M.; Bařinka, C.; Haberkorn, U.; Babich, J. Glu-Ureido–Based Inhibitors of Prostate-Specific Membrane Antigen: Lessons Learned during the Development of a Novel Class of Low-Molecular-Weight Theranostic Radiotracers. J. Nucl. Med. 2017, 58, 17S–26S. [Google Scholar] [CrossRef]
- Kelly, J.; Amor-Coarasa, A.; Ponnala, S.; Nikolopoulou, A.; Williams, C.; Schlyer, D.; Zhao, Y.; Kim, D.; Babich, J.W. Trifunctional PSMA-targeting constructs for prostate cancer with unprecedented localization to LNCaP tumors. Eur. J. Nucl. Med. Mol. Imaging 2018, 45, 1841–1851. [Google Scholar] [CrossRef] [PubMed]
- Kelly, J.M.; Amor-Coarasa, A.; Ponnala, S.; Nikolopoulou, A.; Williams, C.; DiMagno, S.G.; Babich, J. Albumin-Binding PSMA Ligands: Implications for Expanding the Therapeutic Window. J. Nucl. Med. 2019, 60, 656–663. [Google Scholar] [CrossRef]
- Wang, G.; Zang, J.; Jiang, Y.; Liu, Q.; Sui, H.; Wang, R.; Fan, X.; Zhang, J.; Zhu, Z.; Chen, X. A single-arm, low-dose, prospective study of 177Lu-EB-PSMA radioligand therapy in patients with metastatic castration-resistant prostate cancer. J. Nucl. Med. 2022, 64, 611–617. [Google Scholar] [CrossRef] [PubMed]
- Murce, E.; Beekman, S.; Spaan, E.; Handula, M.; Stuurman, D.; de Ridder, C.; Seimbille, Y. Preclinical Evaluation of a PSMA-Targeting Homodimer with an Optimized Linker for Imaging of Prostate Cancer. Molecules 2023, 28, 4022. [Google Scholar] [CrossRef] [PubMed]
- Choy, C.J.; Ling, X.; Geruntho, J.J.; Beyer, S.K.; Latoche, J.D.; Langton-Webster, B.; Anderson, C.J.; Berkman, C.E. 177Lu-Labeled Phosphoramidate-Based PSMA Inhibitors: The Effect of an Albumin Binder on Biodistribution and Therapeutic Efficacy in Prostate Tumor-Bearing Mice. Theranostics 2017, 7, 1928–1939. [Google Scholar] [CrossRef] [PubMed]
- Benešová, M.; Umbricht, C.A.; Schibli, R.; Müller, C. Albumin-Binding PSMA Ligands: Optimization of the Tissue Distribution Profile. Mol. Pharm. 2018, 15, 934–946. [Google Scholar] [CrossRef] [PubMed]
- Lau, J.; Jacobson, O.; Niu, G.; Lin, K.-S.; Bénard, F.; Chen, X. Bench to Bedside: Albumin Binders for Improved Cancer Radioligand Therapies. Bioconjug Chem. 2019, 30, 487–502. [Google Scholar] [CrossRef]
- Zang, J.; Liu, Q.; Sui, H.; Wang, R.; Jacobson, O.; Fan, X.; Zhu, Z.; Chen, X. 177Lu-EB-PSMA Radioligand Therapy with Escalating Doses in Patients with Metastatic Castration-Resistant Prostate Cancer. J. Nucl. Med. 2020, 61, 1772–1778. [Google Scholar] [CrossRef]
- Ling, X.; Latoche, J.D.; Choy, C.J.; Kurland, B.F.; Laymon, C.M.; Wu, Y.; Salamacha, N.; Shen, D.; Geruntho, J.J.; Rigatti, L.H.; et al. Preclinical Dosimetry, Imaging, and Targeted Radionuclide Therapy Studies of Lu-177-Labeled Albumin-Binding, PSMA-Targeted CTT1403. Mol. Imaging Biol. 2020, 22, 274–284. [Google Scholar] [CrossRef]
- Kuo, H.-T.; Lin, K.-S.; Zhang, Z.; Uribe, C.F.; Merkens, H.; Zhang, C.; Bénard, F. 177Lu-Labeled Albumin-Binder–Conjugated PSMA-Targeting Agents with Extremely High Tumor Uptake and Enhanced Tumor-to-Kidney Absorbed Dose Ratio. J. Nucl. Med. 2021, 62, 521–527. [Google Scholar] [CrossRef]
- Reissig, F.; Zarschler, K.; Novy, Z.; Petrik, M.; Bendova, K.; Kurfurstova, D.; Bouchal, J.; Ludik, M.-C.; Brandt, F.; Kopka, K.; et al. Modulating the pharmacokinetic profile of Actinium-225-labeled macropa-derived radioconjugates by dual targeting of PSMA and albumin. Theranostics 2022, 12, 7203–7215. [Google Scholar] [CrossRef] [PubMed]
- Zorzi, A.; Linciano, S.; Angelini, A. Non-covalent albumin-binding ligands for extending the circulating half-life of small biotherapeutics. MedChemComm 2019, 10, 1068–1081. [Google Scholar] [CrossRef] [PubMed]
- Kramer, V.; Fernández, R.; Lehnert, W.; Jiménez-Franco, L.D.; Soza-Ried, C.; Eppard, E.; Ceballos, M.; Meckel, M.; Benešová, M.; Umbricht, C.A.; et al. Biodistribution and dosimetry of a single dose of albumin-binding ligand [177Lu]Lu-PSMA-ALB-56 in patients with mCRPC. Eur. J. Nucl. Med. Mol. Imaging 2021, 48, 893–903. [Google Scholar] [CrossRef] [PubMed]
- Tschan, V.J.; Borgna, F.; Busslinger, S.D.; Stirn, M.; Rodriguez, J.M.M.; Bernhardt, P.; Schibli, R.; Müller, C. Preclinical investigations using [177Lu]Lu-Ibu-DAB-PSMA toward its clinical translation for radioligand therapy of prostate cancer. Eur. J. Nucl. Med. Mol. Imaging 2022, 49, 3639–3650. [Google Scholar] [CrossRef]
- Kuo, H.-T.; Lin, K.-S.; Zhang, Z.; Zhang, C.; Merkens, H.; Tan, R.; Roxin, A.; Uribe, C.F.; Bénard, F. What a difference a methylene makes: Replacing Glu with Asp or Aad in the Lys-urea-Glu pharmacophore of PSMA-targeting radioligands to reduce kidney and salivary gland uptake. Theranostics 2022, 12, 6179–6188. [Google Scholar] [CrossRef]
- Deberle, L.M.; Benešová, M.; Umbricht, C.A.; Borgna, F.; Büchler, M.; Zhernosekov, K.; Schibli, R.; Müller, C. Development of a new class of PSMA radioligands comprising ibuprofen as an albumin-binding entity. Theranostics 2020, 10, 1678–1693. [Google Scholar] [CrossRef]
- Banerjee, S.R.; Kumar, V.; Lisok, A.; Chen, J.; Minn, I.; Brummet, M.; Boinapally, S.; Cole, M.; Ngen, E.; Wharram, B.; et al. 177Lu-labeled low-molecular-weight agents for PSMA-targeted radiopharmaceutical therapy. Eur. J. Nucl. Med. Mol. Imaging 2019, 46, 2545–2557. [Google Scholar] [CrossRef]
- Banerjee, S.R.; Minn, I.; Kumar, V.; Josefsson, A.; Lisok, A.; Brummet, M.; Chen, J.; Kiess, A.P.; Baidoo, K.; Brayton, C.; et al. Preclinical Evaluation of 203/212Pb-Labeled Low-Molecular-Weight Compounds for Targeted Radiopharmaceutical Therapy of Prostate Cancer. J. Nucl. Med. 2020, 61, 80–88. [Google Scholar] [CrossRef]
- Banerjee, S.R.; Lisok, A.; Minn, I.; Josefsson, A.; Kumar, V.; Brummet, M.; Boinapally, S.; Brayton, C.; Mease, R.C.; Sgouros, G.; et al. Preclinical evaluation of 213Bi- and 225Ac-Labeled Low-Molecular-Weight Compounds for Radiopharmaceutical Therapy of Prostate Cancer. J. Nucl. Med. 2021, 62, 980–988. [Google Scholar] [CrossRef]
- Mease, R.C.; Kang, C.M.; Kumar, V.; Banerjee, S.R.; Minn, I.; Brummet, M.; Gabrielson, K.L.; Feng, Y.; Park, A.; Kiess, A.P.; et al. An Improved 211At-Labeled Agent for PSMA-Targeted alpha-Therapy. J. Nucl. Med. 2022, 63, 259–267. [Google Scholar] [CrossRef]
- Olszewski, R.T.; Bukhari, N.; Zhou, J.; Kozikowski, A.P.; Wroblewski, J.T.; Shamimi-Noori, S.; Wroblewska, B.; Bzdega, T.; Vicini, S.; Barton, F.B.; et al. NAAG peptidase inhibition reduces locomotor activity and some stereotypes in the PCP model of schizophrenia via group II mGluR. J. Neurochem. 2004, 89, 876–885. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.R.; Foss, C.A.; Castanares, M.; Mease, R.C.; Byun, Y.; Fox, J.J.; Hilton, J.; Lupold, S.E.; Kozikowski, A.P.; Pomper, M.G. Synthesis and evaluation of technetium-99m- and rhenium-labeled inhibitors of the prostate-specific membrane antigen (PSMA). J. Med. Chem. 2008, 51, 4504–4517. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.R.; Pullambhatla, M.; Byun, Y.; Nimmagadda, S.; Green, G.; Fox, J.J.; Horti, A.; Mease, R.C.; Pomper, M.G. 68Ga-labeled inhibitors of prostate-specific membrane antigen (PSMA) for imaging prostate cancer. J. Med. Chem. 2010, 53, 5333–5341. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Minn, I.; Rowe, S.P.; Lisok, A.; Chatterjee, S.; Brummet, M.; Banerjee, S.R.; Mease, R.C.; Pomper, M.G. A Series of PSMA-Targeted Near-Infrared Fluorescent Imaging Agents. Biomolecules 2022, 12, 405. [Google Scholar] [CrossRef]
- Bacich, D.J.; Pinto, J.T.; Tong, W.P.; Heston, W.D.W. Cloning, expression, genomic localization, and enzymatic activities of the mouse homolog of prostate-specific membrane antigen/NAALADase/folate hydrolase. Mamm. Genome 2001, 12, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Borgna, F.; Deberle, L.M.; Cohrs, S.; Schibli, R.; Müller, C. Combined Application of Albumin-Binding [177Lu]Lu-PSMA-ALB-56 and Fast-Cleared PSMA Inhibitors: Optimization of the Pharmacokinetics. Mol. Pharm. 2020, 17, 2044–2053. [Google Scholar] [CrossRef]
- Busslinger, S.D.; Tschan, V.J.; Richard, O.K.; Talip, Z.; Schibli, R.; Müller, C. [225Ac]Ac-SibuDAB for Targeted Alpha Therapy of Prostate Cancer: Preclinical Evaluation and Comparison with [225Ac]Ac-PSMA-617. Cancers 2022, 14, 5651. [Google Scholar] [CrossRef]
- Banerjee, S.R.; Kumar, V.; Lisok, A.; Plyku, D.; Novakova, Z.; Brummet, M.; Wharram, B.; Barinka, C.; Hobbs, R.; Pomper, M.G. Evaluation of 111In-DOTA-5D3, a Surrogate SPECT Imaging Agent for Radioimmunotherapy of Prostate-Specific Membrane Antigen. J. Nucl. Med. 2019, 60, 400–406. [Google Scholar] [CrossRef]
- Tschan, V.J.; Borgna, F.; Schibli, R.; Müller, C. Impact of the mouse model and molar amount of injected ligand on the tissue distribution profile of PSMA radioligands. Eur. J. Nucl. Med. Mol. Imaging 2021, 49, 470–480. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.R.; Pullambhatla, M.; Byun, Y.; Nimmagadda, S.; Foss, C.A.; Green, G.; Fox, J.J.; Lupold, S.E.; Mease, R.C.; Pomper, M.G. Sequential SPECT and optical imaging of experimental models of prostate cancer with a dual modality inhibitor of the prostate-specific membrane antigen. Angew. Chem. Int. Ed. 2011, 50, 9167–9170. [Google Scholar] [CrossRef]
- Cheng, Y.; Prusoff, W.H. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 1973, 22, 3099–3108. [Google Scholar] [PubMed]
- Tsuchihashi, S.; Nakashima, K.; Tarumizu, Y.; Ichikawa, H.; Jinda, H.; Watanabe, H.; Ono, M. Development of Novel 111In/225Ac-Labeled Agent Targeting PSMA for Highly Efficient Cancer Radiotheranostics. J. Med. Chem. 2023, 66, 8043–8053. [Google Scholar] [CrossRef] [PubMed]
- Leek, J.; Lench, N.; Maraj, B.; Bailey, A.; Carr, I.M.; Andersen, S.; Cross, J.; Whelan, P.; MacLennan, K.A.; Meredith, D.M.; et al. Prostate-specific membrane antigen: Evidence for the existence of a second related human gene. Br. J. Cancer 1995, 72, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.S.; Reuter, V.E.; Heston, W.D.W.; Bander, N.H.; Grauer, L.S.; Gaudin, P.B. Five Different Anti-Prostate-specific Membrane Antigen (PSMA) Antibodies Confirm PSMA Expression in Tumor-associated Neovasculature1. Cancer Res. 1999, 59, 3192–3198. [Google Scholar]
- Liu, C.; Hasegawa, K.; Russell, S.J.; Sadelain, M.; Peng, K.-W. Prostate-specific membrane antigen retargeted measles virotherapy for the treatment of prostate cancer. Prostate 2009, 69, 1128–1141. [Google Scholar] [CrossRef]
- Franken, N.A.; Rodermond, H.M.; Stap, J.; Haveman, J.; van Bree, C. Clonogenic assay of cells in vitro. Nat. Protoc. 2006, 1, 2315–2319. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boinapally, S.; Alati, S.; Jiang, Z.; Yan, Y.; Lisok, A.; Singh, R.; Lofland, G.; Minn, I.; Hobbs, R.F.; Pomper, M.G.; et al. Preclinical Evaluation of a New Series of Albumin-Binding 177Lu-Labeled PSMA-Based Low-Molecular-Weight Radiotherapeutics. Molecules 2023, 28, 6158. https://doi.org/10.3390/molecules28166158
Boinapally S, Alati S, Jiang Z, Yan Y, Lisok A, Singh R, Lofland G, Minn I, Hobbs RF, Pomper MG, et al. Preclinical Evaluation of a New Series of Albumin-Binding 177Lu-Labeled PSMA-Based Low-Molecular-Weight Radiotherapeutics. Molecules. 2023; 28(16):6158. https://doi.org/10.3390/molecules28166158
Chicago/Turabian StyleBoinapally, Srikanth, Suresh Alati, Zirui Jiang, Yu Yan, Alla Lisok, Rajan Singh, Gabriela Lofland, Il Minn, Robert F. Hobbs, Martin G. Pomper, and et al. 2023. "Preclinical Evaluation of a New Series of Albumin-Binding 177Lu-Labeled PSMA-Based Low-Molecular-Weight Radiotherapeutics" Molecules 28, no. 16: 6158. https://doi.org/10.3390/molecules28166158




