Isolation and Characterization of a ssDNA Aptamer against Major Soluble Antigen of Renibacterium salmoninarum
Abstract
:1. Introduction
2. Results and Discussion
2.1. MSA Protein and Constructs Generation
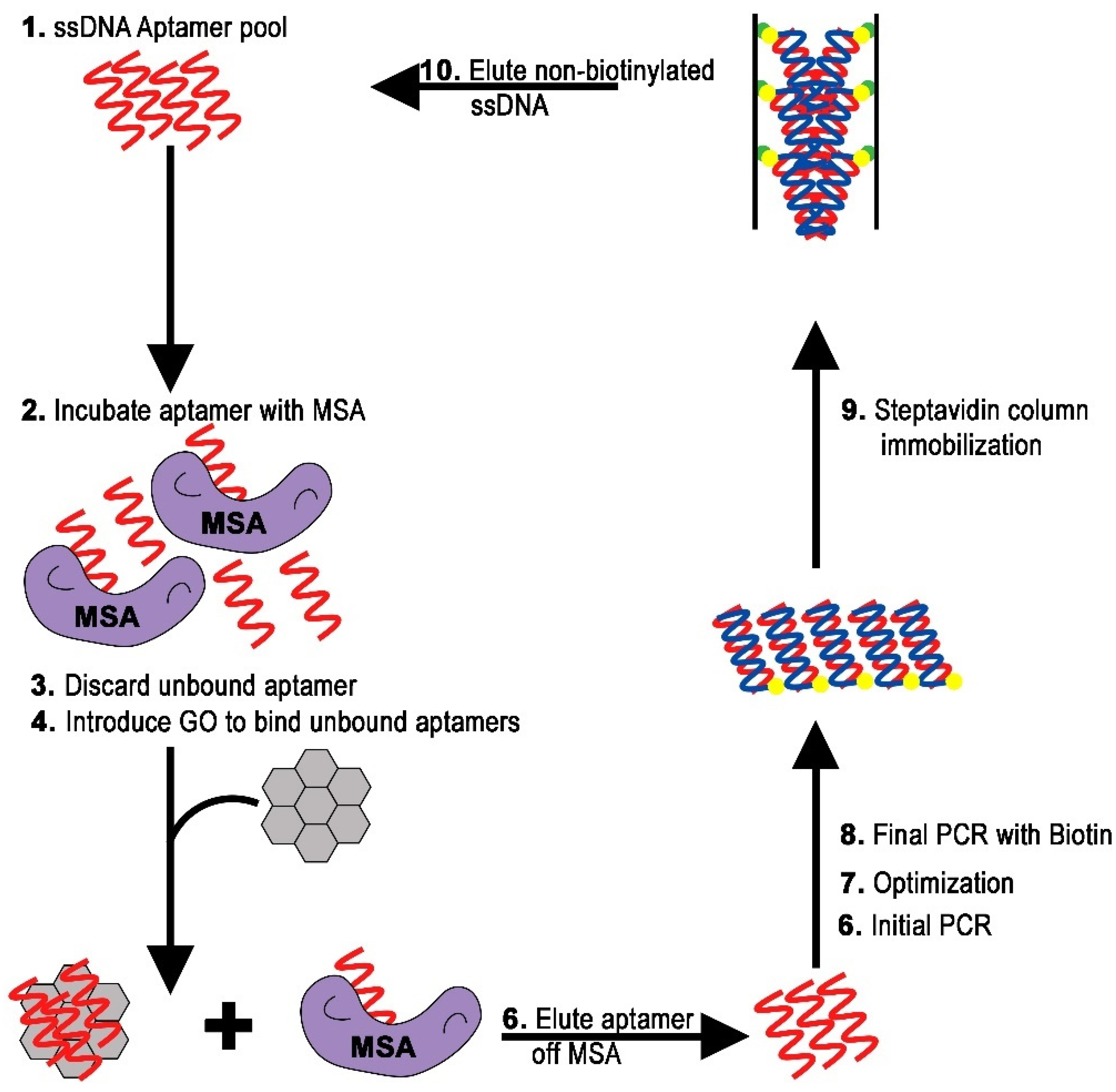
2.2. Isolation of DNA Aptamer
2.3. Analysis of Aptamer Structures
2.4. Binding Affinity and Specificity Test
3. Materials and Methods
3.1. Reagents and Instrumentation
3.2. Cloning, Expression and Purification of MSA Protein and Sub Constructs
3.3. Random Oligonucleotide Library and Primers
3.4. In Vitro Selection via GO-SELEX
3.5. Cloning, Sequencing, and Structure Analysis of Selected Aptamers
3.6. Binding Assay Procedure
3.7. Specificity Test
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fryer, J.L.; Sanders, J.E. Bacterial kidney disease of salmonid fish. Annu. Rev. Microbiol. 1981, 35, 273–298. [Google Scholar] [CrossRef] [PubMed]
- Delghandi, M.R.; El-Matbouli, M.; Menanteau-Ledouble, S. Renibacterium salmoninarum—The Causative Agent of Bacterial Kidney Disease in Salmonid Fish. Pathogens 2020, 9, 845. [Google Scholar] [CrossRef] [PubMed]
- Pascho, R.J.; Goodrich, T.D.; McKibben, C.L. Evaluation by enzyme-linked immunosorbent assay (ELISA) of Renibacterium salmoninarum bacterins affected by persistence of bacterial antigens. J. Aquatic Anim. Health 1997, 9, 99–107. [Google Scholar] [CrossRef]
- Jansson, E.; Lindberg, L.; Saker, E.; Aspan, A. Diagnosis of bacterial kidney disease by detection of Renibacterium salmoninarum by real-time PCR. J. Fish Dis. 2008, 31, 755–763. [Google Scholar] [CrossRef] [PubMed]
- Jansson, E.; Hongslo, T.; Hoglund, J.; Ljungberg, O. Comparative evaluation of bacterial culture and two ELISA techniques for the detection of Renibacterium salmoninarum antigens in salmonid kidney tissues. Dis. Aquat. Org. 1996, 27, 197–206. [Google Scholar] [CrossRef]
- Zhou, J.; Rossi, J. Aptamers as targeted therapeutics: Current potential and challenges. Nat. Rev. Drug Dis. 2017, 16, 181–202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.; Zhou, X.; Ding, W.; Guo, S.; Wu, N. Fluorescent aptamer-functionalized graphene oxide biosensor for label-free detection of mercury (II). Biosens. Bioelectron. 2013, 41, 889–893. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X. Structure-Switching Electrochemical Aptasensor for Single-Step and Specific Detection of Trace Mercury in Dairy Products. J. Agric. Food Chem. 2018, 66, 10106–10112. [Google Scholar] [CrossRef] [PubMed]
- Haixiang, Y.; Luo, Y.; Alkhamis, O.; Canours, J.; Yu, B.; Xiao, Y. Isolation of natural DNA aptamers for challenging small-molecules targets, cannabinoids. Anal. Chem. 2021, 93, 3172–3180. [Google Scholar]
- Li, F.; Zhang, H.; Wang, Z.; Newbigging, A.M.; Reid, M.S.; Li, X.; Le, X.C. Aptamers Facilitating Amplified Detection of Biomolecules. Anal. Chem. 2015, 87, 274–292. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Wu, C.; Tan, W. Cell-Specific Aptamer Characterization. In Aptamers Selected by Cell-SELEX for Theranostics; Tan, W., Fang, X., Eds.; Springer: Berlin/Heidelberg, Germany, 2015. [Google Scholar]
- Park, J.W.; Tatavarti, R.; Kim, D.W.; Jung, H.T.; Gu, M.B. Immobilization-free screening of aptamers assisted by graphene oxide. Chem. Commun. 2012, 48, 2071–2073. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Huang, Y.; Duan, N.; Wu, S.; Xia, Y.; Ma, X.; Zhu, C.; Yuan Jiang, Y.; Wang, Z. Screening and identification of DNA aptamers against T-2 toxin assisted by graphene oxide. J. Agric. Food Chem. 2014, 62, 10368–10374. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Goo, N.; Kim, D. Mechanism of DNA adsorption and desorption on graphene oxide. J. Am. Chem. Soc. 2014, 30, 12587–12595. [Google Scholar] [CrossRef] [PubMed]
- Coady, A.M.; Murray, A.L.; Elliott, D.G.; Rhodes, L.D. Both msa genes in Renibacterium salmoninarum are needed for full virulence in bacterial kidney disease. Appl. Environ. Microbiol. 2006, 72, 2672–2678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiens, G.D.; Chien, M.; Winton, J.R.; Kaattari, S.L. Antigenic and functional characterization of p57 produced by Renibacterium salmoninarum. Dis. Aquat Org. 1999, 37, 43–52. [Google Scholar] [CrossRef]
- Chase, D.M.; Elliott, D.G.; Pascho, R.J. Detection and quantification of Renibacterium salmoninarum DNA in salmonid tissues by real-time quantitative polymerase chain reaction analysis. J. Vet. Diagn. Investig. 2006, 18, 375–380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, A.-Y.; Ha, N.; Jung, I.; Kim, S.; Kim, A.; Yoon, M. Development of a ssDNA aptamer for detection of residual benzypenicillin. Analyt. Biochem. 2017, 531, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Ranganathan, S.V.; Halvorsen, K.; Myers, C.A.; Robertson, N.M.; Yigit, M.V.; Chen, A.A. Complex thermodynamic behavior of single-stranded nucleic acid adsorption to graphene surfaces. J. Am. Chem. Soc. 2016, 32, 6028–6034. [Google Scholar] [CrossRef] [PubMed]
- Kong, B.S.; Geng, J.X.; Jung, H.T. Layer-by-layer assembly of graphene and gold nanoparticles by vacuum filtration and spontaneous reduction of gold ions. Chem. Commun. 2009, 16, 2174–2176. [Google Scholar] [CrossRef]
- Marimuthu, C.; Tang, T.; Tominaga, J.; Tan, S.; Gopinath, S. Single-stranded DNA (ssDNA) production in DNA aptamer generation. Analyst 2012, 137, 1307–1315. [Google Scholar] [CrossRef]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]
- Sano, T.; Smith, C.L.; Cantor, C.R. Immuno-PCR: Very sensitive antigen detection by means of specific antibody-DNA conjugates. Science 1992, 258, 120–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Avci-Adali, M.; Wilhelm, N.; Perle, N.; Stoll, H.; Schlensak, C.; Wendel, H.P. Absolute Quantification of Cell-Bound DNA Aptamers During SELEX. Nucleic Acid Ther. 2013, 23, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, S.; Yuan, J. Binding Characteristics Study of DNA based Aptamers for E. coli O157:H7. Molecules 2021, 26, 204. [Google Scholar] [CrossRef] [PubMed]
- Moreno, M.; Fernández-Algar, M.; Fernández-Chamorro, J.; Ramajo, J.; Martínez-Salas, E.; Briones, C. A Combined ELONA-(RT)qPCR Approach for Characterizing DNA and RNA Aptamers Selected against PCBP-2. Molecules 2019, 24, 1213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jing, M.; Bowser, M. A Review of Methods for Measuring Aptamer-Protein Equilibria. Anal. Chim. Acta 2011, 686, 9–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Name | DNA Aptamer Sequence | Frequency |
|---|---|---|
| Aptamer-1 | 5′-ATGATACGGCGACCACCTAACGCACACAATATATTAAGGATCAAATTAATACTCGGTATATAGCGTCGTGTAGGGAAAGA-3′ | 1 |
| Aptamer-2 | 5′-ATGATACGGCGACCACCTAAACATATGGAGTATTAGGGTTTACATAATTATGTATTTCTCTAGCGTCGTGTAGGGAAAGA-3′ | 1 |
| Aptamer-3 | 5′-ATGATACGGCGACCACCTAAATTACTACATTAATTAGTTGTATAATTGTAGTATGATGATTAGCGTCGTGTAGGGAAAGA-3′ | 2 |
| Aptamer-4 | 5′-ATGATACGGCGACCACCTAACCATTTTCACTGTTTGTTTAATATATATAGTTTTATGGTATAGCGTCGTGTAGGGAAAGA-3′ | 6 |
| Aptamer-5 | 5′-ATGATACGGCGACCACCTAAAGTTATTTTTGTTATTATAACGGGTTACCTTTATAGACTATTAGCGTCGTGTAGGGAAAGA-3′ | 1 |
| Aptamer-6 | 5′-ATGATACGGCGACCACCTAATTTATTAACTATTAACTGTTTAGGTGAGACTTTTCTGTTGTAGCGTCGTGTAGGGAAAGA-3′ | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Layman, B.; Mandella, B.; Carter, J.; Breen, H.; Rinehart, J.; Cavinato, A. Isolation and Characterization of a ssDNA Aptamer against Major Soluble Antigen of Renibacterium salmoninarum. Molecules 2022, 27, 1853. https://doi.org/10.3390/molecules27061853
Layman B, Mandella B, Carter J, Breen H, Rinehart J, Cavinato A. Isolation and Characterization of a ssDNA Aptamer against Major Soluble Antigen of Renibacterium salmoninarum. Molecules. 2022; 27(6):1853. https://doi.org/10.3390/molecules27061853
Chicago/Turabian StyleLayman, Brady, Brian Mandella, Jessica Carter, Haley Breen, John Rinehart, and Anna Cavinato. 2022. "Isolation and Characterization of a ssDNA Aptamer against Major Soluble Antigen of Renibacterium salmoninarum" Molecules 27, no. 6: 1853. https://doi.org/10.3390/molecules27061853





