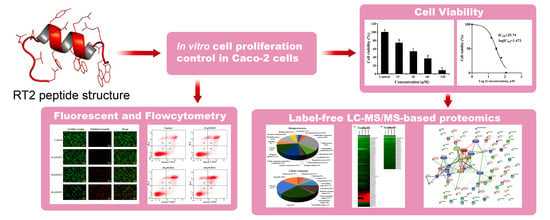
An Integrated Proteomics and Bioinformatics Analysis of the Anticancer Properties of RT2 Antimicrobial Peptide on Human Colon Cancer (Caco-2) Cells
Abstract
:1. Introduction
2. Results and Discussion
2.1. Cytotoxic Effect of RT2 on Caco-2 Cells
2.2. Caco-2 Cell Label-Free Based Proteomics Profiles
2.3. Caco-2 Cell Proteomics Profiling Based on the Percentage of Cell Viability
2.4. Apoptotic Effect of RT2 on Caco-2 Cells
2.5. Caco-2 Cell Proteomics Profiling Based on Apoptosis Level
2.6. mRNA Expression Levels of Genes Related to Cell Viability and Apoptosis Level
3. Materials and Methods
3.1. RT2 Peptide Synthesis
3.2. Cell Culture
3.3. MTT Caco-2 Cell Proliferation and Cytotoxicity Assay
3.4. Total Protein Extraction for Proteomic Assay
3.5. In-Solution Trypsin Digestion for Proteomic Assay
3.6. Label-Free LC-MS/MS-Based Proteomics
3.7. Protein Quantitation and Identification
3.8. Proteomics Data Bioinformatics Analysis
3.9. Apoptosis Fluorescence Morphological Assay
3.10. Apoptosis Flow Cytometric Assay
3.11. RT-qPCR Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Gabernet, G.; Gautschi, D.; Müller, A.T.; Neuhaus, C.S.; Armbrecht, L.; Dittrich, P.S.; Hiss, J.A.; Schneider, G. In silico design and optimization of selective membranolytic anticancer peptides. Sci. Rep. 2019, 9, 11282. [Google Scholar] [CrossRef]
- Phi, L.T.H.; Sari, I.N.; Yang, Y.-G.; Lee, S.-H.; Jun, N.; Kim, K.S.; Lee, Y.K.; Kwon, H.Y. Cancer stem cells (CSCs) in drug resistance and their therapeutic implications in cancer treatment. Stem Cells Int. 2018, 2018, 5416923. [Google Scholar] [CrossRef] [Green Version]
- Housman, G.; Byler, S.; Heerboth, S.; Lapinska, K.; Longacre, M.; Snyder, N.; Sarkar, S. Drug resistance in cancer: An overview. Cancers 2014, 6, 1769–1792. [Google Scholar] [CrossRef] [Green Version]
- Mouhid, L.; Corzo-Martinez, M.; Torres, C.; Vazquez, L.; Reglero, G.; Fornari, T.; Ramirez de Molina, A. Improving in vivo efficacy of bioactive molecules: An overview of potentially antitumor phytochemicals and currently available lipid-based delivery systems. J. Oncol. 2017, 2017, 7351976. [Google Scholar] [CrossRef] [Green Version]
- Anunthawan, T.; de la Fuente-Núñez, C.; Hancock, R.E.W.; Klaynongsruang, S. Cationic amphipathic peptides KT2 and RT2 are taken up into bacterial cells and kill planktonic and biofilm bacteria. Biochim. Biophys. Acta 2015, 1848, 1352–1358. [Google Scholar] [CrossRef] [Green Version]
- Lei, J.; Sun, L.; Huang, S.; Zhu, C.; Li, P.; He, J.; Mackey, V.; Coy, D.H.; He, Q. The antimicrobial peptides and their potential clinical applications. Am. J. Transl. Res. 2019, 11, 3919–3931. [Google Scholar]
- Zhang, S.K.; Ma, Q.; Li, S.B.; Gao, H.W.; Tan, Y.X.; Gong, F.; Ji, S.P. RV-23, a melittin-related peptide with cell-selective antibacterial activity and high hemocompatibility. J. Microbiol. Biotechnol. 2016, 26, 1046–1056. [Google Scholar] [CrossRef] [Green Version]
- Sang, M.; Zhang, J.; Zhuge, Q. Selective cytotoxicity of the antibacterial peptide ABP-dHC-Cecropin A and its analog towards leukemia cells. Eur. J. Pharmacol. 2017, 803, 138–147. [Google Scholar] [CrossRef]
- Xu, P.; Lv, D.; Wang, X.; Wang, Y.; Hou, C.; Gao, K.; Guo, X. Inhibitory effects of Bombyx mori antimicrobial peptide cecropins on esophageal cancer cells. Eur. J. Pharmacol. 2020, 887, 173434. [Google Scholar] [CrossRef]
- Ahmad, A.; Ahmad, E.; Rabbani, G.; Haque, S.; Arshad, M.; Khan, R.H. Identification and design of antimicrobial peptides for therapeutic applications. Curr. Protein Pept. Sci. 2012, 13, 211–223. [Google Scholar] [CrossRef]
- Theansungnoen, T.; Maijaroen, S.; Jangpromma, N.; Yaraksa, N.; Daduang, S.; Temsiripong, T.; Daduang, J.; Klaynongsruang, S. Cationic antimicrobial peptides derived from Crocodylus siamensis leukocyte extract, revealing anticancer activity and apoptotic induction on human cervical cancer cells. Protein J. 2016, 35, 202–211. [Google Scholar] [CrossRef]
- Payoungkiattikun, W.; Joompang, A.; Thongchot, S.; Nowichai, B.; Jangpromma, N.; Klaynongsruang, S. Evidence of multi-functional peptide activity: Potential role of KT2 and RT2 for anti-inflammatory, anti-oxidative stress, and anti-apoptosis properties. Appl. Biol. Chem. 2020, 63, 5. [Google Scholar] [CrossRef]
- Maijaroen, S.; Jangpromma, N.; Daduang, J.; Klaynongsruang, S. KT2 and RT2 modified antimicrobial peptides derived from Crocodylus siamensis Leucrocin I show activity against human colon cancer HCT-116 cells. Environ. Toxicol. Pharmacol. 2018, 62, 164–176. [Google Scholar] [CrossRef]
- Fesler, A.; Guo, S.; Liu, H.; Wu, N.; Ju, J. Overcoming chemoresistance in cancer stem cells with the help of microRNAs in colorectal cancer. Epigenomics 2017, 9, 793–796. [Google Scholar] [CrossRef]
- Van der Jeught, K.; Xu, H.C.; Li, Y.J.; Lu, X.B.; Ji, G. Drug resistance and new therapies in colorectal cancer. World J. Gastroenterol. 2018, 24, 3834–3848. [Google Scholar] [CrossRef]
- Gaspar, D.; Veiga, A.S.; Castanho, M.A.R.B. From antimicrobial to anticancer peptides. A review. Front. Microbiol. 2013, 4, 294. [Google Scholar] [CrossRef] [Green Version]
- Schweizer, F. Cationic amphiphilic peptides with cancer-selective toxicity. Eur. J. Pharmacol. 2009, 625, 190–194. [Google Scholar] [CrossRef]
- Wang, L.; Huang, Y.; Wang, X.; Chen, Y. Label-free LC-MS/MS proteomics analyses reveal proteomic changes accompanying MSTN KO in C2C12 cells. Biomed. Res. Int. 2019, 2019, 7052456. [Google Scholar] [CrossRef] [Green Version]
- Murray, J.E.; Laurieri, N.; Delgoda, R. Proteins. In Pharmacognosy; Badal, S., Delgoda, R., Eds.; Academic Press: Boston, MA, USA, 2017; pp. 477–494. [Google Scholar]
- Ciccolini, J.; Fina, F.; Bezulier, K.; Giacometti, S.; Roussel, M.; Evrard, A.; Cuq, P.; Romain, S.; Martin, P.-M.; Aubert, C. Transmission of apoptosis in human colorectal tumor cells exposed to capecitabine, xeloda, is mediated via Fas. Mol. Cancer. Ther. 2002, 1, 923–927. [Google Scholar]
- Koedrith, P.; Seo, Y.R. Induction of doxorubicin-mediated apoptosis via thioredoxin reductase 1 RNAi in human colon cancer cells. Mol. Cell. Toxicol. 2011, 7, 112–119. [Google Scholar] [CrossRef]
- Nita, M.E.; Nagawa, H.; Tominaga, O.; Tsuno, N.; Fujii, S.; Sasaki, S.; Fu, C.G.; Takenoue, T.; Tsuruo, T.; Muto, T. 5-Fluorouracil induces apoptosis in human colon cancer cell lines with modulation of Bcl-2 family proteins. Br. J. Cancer 1998, 78, 986–992. [Google Scholar] [CrossRef] [Green Version]
- Tan, S.; Peng, X.; Peng, W.; Zhao, Y.; Wei, Y. Enhancement of oxaliplatin-induced cell apoptosis and tumor suppression by 3-methyladenine in colon cancer. Oncol. Lett. 2015, 9, 2056–2062. [Google Scholar] [CrossRef]
- Liu, R.; Huang, S.; Lei, Y.; Zhang, T.; Wang, K.; Liu, B.; Nice, E.C.; Xiang, R.; Xie, K.; Li, J.; et al. FGF8 promotes colorectal cancer growth and metastasis by activating YAP1. Oncotarget 2015, 6, 935–952. [Google Scholar] [CrossRef] [Green Version]
- Marsh, S.K.; Bansal, G.S.; Zammit, C.; Barnard, R.; Coope, R.; Roberts-Clarke, D.; Gomm, J.J.; Coombes, R.C.; Johnston, C.L. Increased expression of fibroblast growth factor 8 in human breast cancer. Oncogene 1999, 18, 1053–1060. [Google Scholar] [CrossRef] [Green Version]
- Turner, N.; Grose, R. Fibroblast growth factor signalling: From development to cancer. Nat. Rev. Cancer 2010, 10, 116–129. [Google Scholar] [CrossRef]
- Mattila, M.M.; Härkönen, P.L. Role of fibroblast growth factor 8 in growth and progression of hormonal cancer. Cytokine Growth Factor Rev. 2007, 18, 257–266. [Google Scholar] [CrossRef]
- Franzon, V.L.; Gibson, M.A.; Hatzinikolas, G.; Woollatt, E.; Sutherland, G.R.; Cleary, E.G. Molecular cloning of a novel human PAPS synthetase which is differentially expressed in metastatic and non-metastatic colon carcinoma cells. Int. J. Biochem. Cell Biol. 1999, 31, 613–626. [Google Scholar] [CrossRef]
- Zhang, Y.; Zou, X.; Qian, W.; Weng, X.; Zhang, L.; Zhang, L.; Wang, S.; Cao, X.; Ma, L.; Wei, G.; et al. Enhanced PAPSS2/VCAN sulfation axis is essential for Snail-mediated breast cancer cell migration and metastasis. Cell Death Differ. 2019, 26, 565–579. [Google Scholar] [CrossRef] [Green Version]
- Jung, S.H.; Lee, H.C.; Yu, D.M.; Kim, B.C.; Park, S.M.; Lee, Y.S.; Park, H.J.; Ko, Y.G.; Lee, J.S. Heparan sulfation is essential for the prevention of cellular senescence. Cell Death Differ. 2016, 23, 417–429. [Google Scholar] [CrossRef] [Green Version]
- Ji, J.; Zhou, C.; Wu, J.; Cai, Q.; Shi, M.; Zhang, H.; Yu, Y.; Zhu, Z.; Zhang, J. Expression pattern of CDK12 protein in gastric cancer and its positive correlation with CD8(+) cell density and CCL12 expression. Int. J. Med. Sci. 2019, 16, 1142–1148. [Google Scholar] [CrossRef] [Green Version]
- Paculová, H.; Kohoutek, J. The emerging roles of CDK12 in tumorigenesis. Cell Div. 2017, 12, 7. [Google Scholar] [CrossRef] [Green Version]
- Pilarova, K.; Herudek, J.; Blazek, D. CDK12: Cellular functions and therapeutic potential of versatile player in cancer. NAR Cancer 2020, 2, zcaa003. [Google Scholar] [CrossRef] [Green Version]
- Feng, Y.; Xiong, Y.; Qiao, T.; Li, X.; Jia, L.; Han, Y. Lactate dehydrogenase A: A key player in carcinogenesis and potential target in cancer therapy. Cancer Med. 2018, 7, 6124–6136. [Google Scholar] [CrossRef] [Green Version]
- Xian, Z.Y.; Liu, J.M.; Chen, Q.K.; Chen, H.Z.; Ye, C.J.; Xue, J.; Yang, H.Q.; Li, J.L.; Liu, X.F.; Kuang, S.J. Inhibition of LDHA suppresses tumor progression in prostate cancer. Tumour Biol. 2015, 36, 8093–8100. [Google Scholar] [CrossRef] [Green Version]
- Jang, M.; Kim, S.S.; Lee, J. Cancer cell metabolism: Implications for therapeutic targets. Exp. Mol. Med. 2013, 45, e45. [Google Scholar] [CrossRef] [Green Version]
- Koukourakis, M.; Giatromanolaki, A.; Sivridis, E. Colorectal cancer: Lactate dehydrogenase (LDH) activity as a prognostic marker. Colorectal Cancer 2009, 4, 241–253. [Google Scholar]
- Urbańska, K.; Orzechowski, A. Unappreciated role of LDHA and LDHB to control apoptosis and autophagy in tumor cells. Int. J. Mol. Sci. 2019, 20, 2085. [Google Scholar] [CrossRef] [Green Version]
- Khaodee, W.; Udomsom, S.; Kunnaja, P.; Cressey, R. Knockout of glucosidase II beta subunit inhibits growth and metastatic potential of lung cancer cells by inhibiting receptor tyrosine kinase activities. Sci. Rep. 2019, 9, 10394. [Google Scholar] [CrossRef] [Green Version]
- Cui, J.; Chen, B.; Wang, H.; Han, Y.; Chen, X.; Zhang, W. Glucosidase II β-subunit, a novel substrate for caspase-3-like activity in rice, plays as a molecular switch between autophagy and programmed cell death. Sci. Rep. 2016, 6, 31764. [Google Scholar] [CrossRef] [Green Version]
- Shin, G.-C.; Moon, S.U.; Kang, H.S.; Choi, H.-S.; Han, H.D.; Kim, K.-H. PRKCSH contributes to tumorigenesis by selective boosting of IRE1 signaling pathway. Nat. Commun. 2019, 10, 3185. [Google Scholar] [CrossRef] [Green Version]
- Pimiento, J.M.; Neill, K.G.; Henderson-Jackson, E.; Eschrich, S.A.; Chen, D.-T.; Husain, K.; Shibata, D.; Coppola, D.; Malafa, M.P. Knockdown of CSE1L gene in colorectal cancer reduces tumorigenesis in vitro. Am. J. Pathol. 2016, 186, 2761–2768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, J.H.; Hong, D.F.; Song, Y.M.; Sun, L.F.; Wang, Z.F.; Wang, J.W. Suppression of cellular apoptosis susceptibility (CSE1L) inhibits proliferation and induces apoptosis in colorectal cancer cells. Asian Pac. J. Cancer Prev. 2013, 14, 1017–1021. [Google Scholar] [CrossRef] [Green Version]
- Piktel, E.; Niemirowicz, K.; Wnorowska, U.; Wątek, M.; Wollny, T.; Głuszek, K.; Góźdź, S.; Levental, I.; Bucki, R. The role of cathelicidin LL-37 in cancer development. Arch. Immunol. Ther. Exp. 2016, 64, 33–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gajski, G.; Garaj-Vrhovac, V. Melittin: A lytic peptide with anticancer properties. Environ. Toxicol. Pharmacol. 2013, 36, 697–705. [Google Scholar] [CrossRef]
- Kim, S.; Kim, S.S.; Bang, Y.J.; Kim, S.J.; Lee, B.J. In vitro activities of native and designed peptide antibiotics against drug sensitive and resistant tumor cell lines. Peptides 2003, 24, 945–953. [Google Scholar] [CrossRef]
- Nasrallah, A.; Saykali, B.; Al Dimassi, S.; Khoury, N.; Hanna, S.; El-Sibai, M. Effect of StarD13 on colorectal cancer proliferation, motility and invasion. Oncol. Rep. 2014, 31, 505–515. [Google Scholar] [CrossRef] [Green Version]
- Gao, F.; Yu, X.; Meng, R.; Wang, J.; Jia, L. STARD13 is positively correlated with good prognosis and enhances 5-FU sensitivity via suppressing cancer stemness in hepatocellular carcinoma cells. Onco. Targets Ther. 2018, 11, 5371–5381. [Google Scholar] [CrossRef] [Green Version]
- Zhou, G.; Liu, X.; Xiong, B.; Sun, Y. Homeobox B4 inhibits breast cancer cell migration by directly binding to StAR-related lipid transfer domain protein 13. Oncol. Lett. 2017, 14, 4625–4632. [Google Scholar] [CrossRef] [Green Version]
- Peng, L.N.; Deng, X.Y.; Gan, X.X.; Zhang, J.H.; Ren, G.H.; Shen, F.; Feng, J.H.; Cai, W.S.; Xu, B. Targeting of TLE3 by miR-3677 in human breast cancer promotes cell proliferation, migration and invasion. Oncol. Lett. 2020, 19, 1409–1417. [Google Scholar] [CrossRef] [Green Version]
- Yang, R.-W.; Zeng, Y.-Y.; Wei, W.-T.; Cui, Y.-M.; Sun, H.-Y.; Cai, Y.-L.; Nian, X.-X.; Hu, Y.-T.; Quan, Y.-P.; Jiang, S.-L.; et al. TLE3 represses colorectal cancer proliferation by inhibiting MAPK and AKT signaling pathways. J. Eep. Clin. Cancer Res. 2016, 35, 152. [Google Scholar] [CrossRef] [Green Version]
- Paraiso, K.H.; Van Der Kooi, K.; Messina, J.L.; Smalley, K.S. Measurement of constitutive MAPK and PI3K/AKT signaling activity in human cancer cell lines. Methods Enzymol. 2010, 484, 549–567. [Google Scholar]
- Koveitypour, Z.; Panahi, F.; Vakilian, M.; Peymani, M.; Seyed Forootan, F.; Nasr Esfahani, M.H.; Ghaedi, K. Signaling pathways involved in colorectal cancer progression. Cell Biosci. 2019, 9, 97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.; Zhang, Y.; Wong, C.C.; Zhang, J.; Dong, Y.; Li, X.; Kang, W.; Chan, F.K.L.; Sung, J.J.Y.; Yu, J. RNF6 promotes colorectal cancer by activating the Wnt/β-catenin pathway via ubiquitination of TLE3. Cancer Res. 2018, 78, 1958–1971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fedorova, M.S.; Kudryavtseva, A.V.; Lakunina, V.A.; Snezhkina, A.V.; Volchenko, N.N.; Slavnova, E.N.; Danilova, T.V.; Sadritdinova, A.F.; Melnikova, N.V.; Belova, A.A.; et al. Downregulation of OGDHL expression is associated with promoter hypermethylation in colorectal cancer. Mol. Biol. 2015, 49, 678–688. [Google Scholar] [CrossRef]
- Sen, T.; Sen, N.; Noordhuis, M.G.; Ravi, R.; Wu, T.C.; Ha, P.K.; Sidransky, D.; Hoque, M.O. OGDHL is a modifier of AKT-dependent signaling and NF-κB function. PLoS ONE 2012, 7, e48770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anunthawan, T.; Yaraksa, N.; Phosri, S.; Theansungnoen, T.; Daduang, S.; Dhiravisit, A.; Thammasirirak, S. Improving the antibacterial activity and selectivity of an ultra short peptide by hydrophobic and hydrophilic amino acid stretches. Bioorg. Med. Chem. Lett. 2013, 23, 4657–4662. [Google Scholar] [CrossRef]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Thorsell, A.; Portelius, E.; Blennow, K.; Westman-Brinkmalm, A. Evaluation of sample fractionation using micro-scale liquid-phase isoelectric focusing on mass spectrometric identification and quantitation of proteins in a SILAC experiment. Rapid Commun Mass Spectrom. 2007, 21, 771–778. [Google Scholar] [CrossRef]
- Perkins, D.N.; Pappin, D.J.; Creasy, D.M.; Cottrell, J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 1999, 20, 3551–3567. [Google Scholar] [CrossRef]
- Howe, E.; Holton, K.; Nair, S.; Schlauch, D.; Sinha, R.; Quackenbush, J. MeV: MultiExperiment viewer. In Biomedical Informatics for Cancer Research; Ochs, M.F., Casagrande, J.T., Davuluri, R.V., Eds.; Springer: Boston, MA, USA, 2010; pp. 267–277. [Google Scholar]
- Wang, Y.; Zhang, Y.; Zhou, R.; Dossa, K.; Yu, J.; Li, D.; Liu, A.; Mmadi, M.A.; Zhang, X.; You, J. Identification and characterization of the bZIP transcription factor family and its expression in response to abiotic stresses in sesame. PLoS ONE 2018, 13, e0200850. [Google Scholar] [CrossRef]
- Mi, H.; Huang, X.; Muruganujan, A.; Tang, H.; Mills, C.; Kang, D.; Thomas, P.D. PANTHER version 11: Expanded annotation data from gene ontology and reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017, 45, 183–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szklarczyk, D.; Santos, A.; von Mering, C.; Jensen, L.J.; Bork, P.; Kuhn, M. STITCH 5: Augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 2016, 44, 380–384. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]








| Accession | Protein Name | Gene Name | Anova (p) | q Value | Treatment Effect (Fold Change) | ||
|---|---|---|---|---|---|---|---|
| Caco-2 Treated with 15 µM RT2 Compared to Untreated Control | Caco-2 Treated with 30 µM RT2 Compared to Untreated Control | Caco-2 Treated with 60 µM RT2 Compared to Untreated Control | |||||
| P55075.1 | Fibroblast growth factor 8 (FGF-8) | FGF8 | 0.02300 | 0.00137 | +1.03 | −1.23 | −1.11 |
| O95340.2 | Bifunctional 3′-phosphoadenosine 5′-phosphosulfate synthase 2 | PAPSS2 | 0.05516 | 0.00233 | −1.00 | −1.17 | −1.32 |
| Q9NYV4.2 | Cyclin-dependent kinase 12 | CDK12 | 0.07786 | 0.00298 | −1.05 | −1.10 | −1.15 |
| P00338.2 | L-lactate dehydrogenase A chain | LDHA | 0.00001 | 0.00000 | +1.15 | −1.25 | −1.18 |
| P14314.2 | Glucosidase 2 subunit beta (80K-H protein) | PRKCSH | 0.00128 | 0.00018 | −1.24 | −1.23 | −1.74 |
| P55060.3 | Exportin-2 (Exp2) (Cellular apoptosis susceptibility protein) | CSE1L | 0.00395 | 0.00043 | +1.06 | −1.08 | −1.14 |
| Q9Y6X0.3 | SET-binding protein (SEB) | SETBP1 | 0.00002 | 0.00001 | −1.08 | −1.15 | −1.33 |
| O95239.3 | Chromosome-associated kinesin KIF4A (Chromokinesin-A) | KIF4A4 | 0.00600 | 0.00056 | −1.10 | −1.19 | −1.33 |
| P62258.1 | 14-3-3 protein epsilon (14-3-3E) | YWHAE | 0.00190 | 0.00024 | −1.09 | −1.12 | −1.18 |
| Q969U6.1 | F-box/WD repeat-containing protein 5 | FBXW5 | 0.13075 | 0.00436 | +1.03 | −1.10 | −1.05 |
| P0CG47.1 | Polyubiquitin-B [Cleaved into: Ubiquitin] | UBB | 0.00432 | 0.00046 | −1.09 | −1.17 | −1.24 |
| Q15788.3 | Nuclear receptor coactivator 1 (NCoA-1) | NCOA1 | 0.00007 | 0.00002 | +1.14 | −1.39 | −1.21 |
| O14646.2 | Chromodomain-helicase-DNA-binding protein 1 | CHD1 | 0.00843 | 0.00072 | −1.14 | −1.11 | −1.28 |
| Q92766.3 | Ras-responsive element-binding protein 1 | RREB1 | 0.00040 | 0.00008 | −1.05 | −1.13 | −1.18 |
| P60174.3 | Triosephosphate isomerase | TPI1 | 0.00639 | 0.00059 | −1.04 | −1.16 | −1.20 |
| Q96R06.2 | Sperm-associated antigen 5 (Astrin) | SPAG5 | 0.00599 | 0.00056 | −1.07 | −1.11 | −1.17 |
| O15229.2 | Kynurenine 3-monooxygenase | KMO | 0.00882 | 0.00074 | −1.08 | −1.09 | −1.16 |
| O43707.2 | Alpha-actinin-4 (Non-muscle alpha-actinin 4) | ACTN4 | 0.00265 | 0.00031 | +1.01 | −1.13 | −1.11 |
| Q9H2K2.1 | Poly [ADP-ribose] polymerase tankyrase-2 | TNKS2 | 0.37048 | 0.01071 | −1.05 | −1.08 | −1.09 |
| Q9Y4B5.5 | Microtubule cross-linking factor 1 | MTCL1 | 0.05532 | 0.00234 | −1.05 | −1.09 | −1.15 |
| Accession | Protein Name | Gene Name | Anova (p) | q Value | Treatment Effect (Fold Change) | ||
|---|---|---|---|---|---|---|---|
| Caco-2 Treated with 15 µM RT2 Compared to Untreated Control | Caco-2 Treated with 30 µM RT2 Compared to Untreated Control | Caco-2 Treated with 60 µM RT2 Compared to Untreated Control | |||||
| Q9Y3M8.2 | StAR-related lipid transfer protein 13 | STARD13 | 0.01026 | 0.00083 | +1.03 | +1.51 | +1.38 |
| Q9ULD0.3 | 2-oxoglutarate dehydrogenase-like, mitochondrial | OGDHL | 0.00001 | 0.00000 | −1.02 | +1.25 | +2.20 |
| Q04726.2 | Transducin-like enhancer protein 3 | TLE3 | 0.00093 | 0.00014 | −1.00 | +1.14 | +1.27 |
| O75140.2 | GATOR complex protein DEPDC5 | DEPDC5 | 0.05765 | 0.00239 | +1.07 | +1.10 | +1.21 |
| P86452.1 | Zinc finger BED domain-containing protein 6 | ZBED6 | 0.00000 | 0.00000 | +1.05 | +1.41 | +3.18 |
| O43143.2 | Pre-mRNA-splicing factor ATP-dependent RNA helicase | DHX15 | 0.00000 | 0.00000 | +1.19 | +1.74 | +3.90 |
| Q9HBU1.2 | Homeobox protein BarH-like 1 | BARX1 | 0.00000 | 0.00000 | +1.05 | +1.32 | +2.15 |
| Q8TE57.3 | A disintegrin and metalloproteinase with thrombospondin motifs 16 | ADAMTS16 | 0.00173 | 0.00023 | −1.03 | +1.27 | +1.45 |
| Q13796.1 | Protein Shroom2 (Apical-like protein) (Protein APXL) | SHROOM2 | 0.00000 | 0.00000 | +1.06 | +1.06 | +1.58 |
| Q5T5X7.1 | BEN domain-containing protein 3 | BEND3 | 0.15843 | 0.00510 | +1.10 | +1.22 | +1.06 |
| O95803.1 | Bifunctional heparan sulfate N-deacetylase/ N-sulfotransferase 3 | NDST3 | 0.39396 | 0.01136 | +1.03 | +1.08 | +1.04 |
| Gene | Primer Sequence (5′→3′) | Annealing | Product | Reference |
|---|---|---|---|---|
| (°C) | Size (bp) | Sequence | ||
| FGF8 | F_ TGTCTCCCAACAGCATGTGA | 59 | 181 | NM_033164.4 |
| R_ CGTCTCCACGATGAGCTTTG | ||||
| PAPSS2 | F_ CAAACTTGACCACGTCCGAG | 57 | 179 | NM_004670.4 |
| R_ TCACGCCATCATCTAGCAGG | ||||
| CDK12 | F_ AGTCCACTCCCCAGTAGGAA | 57 | 168 | NM_016507.4 |
| R_ ACTGAGTTCAGCTCCCAGAC | ||||
| LDHA | F_ GAACAGTGGAAAGAGTGCAGAT | 57 | 132 | NM_001165415.2 |
| R_ AGGACAACATGCACAACCTC | ||||
| PRKCSH | F_ CACAATTGGGCACAGGGAG | 59 | 129 | NM_001289102.1 |
| R_ GGCCTCACTTTGTTTCCGAT | ||||
| CSE1L | F_ GGCACAGTCACTTCACAAGTT | 58 | 171 | NM_001362762.2 |
| R_ CGCCAATACAAACCCCATCTT | ||||
| STRAD13 | F_ GCCGAGATGTTCAGTCAGGT | 57 | 159 | NM_178006.4 |
| R_ CACTAGCTGATGGCGTGCTA | ||||
| OGDHL | F_ GTTTCTTCAAACGTGGATCTTGC | 60 | 139 | NM_001363523.2 |
| R_ ATTCCTGTCCCCCGATGAAA | ||||
| TLE3 | F_ CTCGCCCTTGTCAGCTCTTA | 57 | 200 | NM_001282982.2 |
| R_ GTTCAGCTCCGTCATGGTGA | ||||
| GAPDH | F_ GAAATGAATGGGCAGCCGTT | 59 | 173 | NM_001256799.3 |
| R_ CGCCCAATACGACCAAATCAG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maijaroen, S.; Klaynongsruang, S.; Roytrakul, S.; Konkchaiyaphum, M.; Taemaitree, L.; Jangpromma, N. An Integrated Proteomics and Bioinformatics Analysis of the Anticancer Properties of RT2 Antimicrobial Peptide on Human Colon Cancer (Caco-2) Cells. Molecules 2022, 27, 1426. https://doi.org/10.3390/molecules27041426
Maijaroen S, Klaynongsruang S, Roytrakul S, Konkchaiyaphum M, Taemaitree L, Jangpromma N. An Integrated Proteomics and Bioinformatics Analysis of the Anticancer Properties of RT2 Antimicrobial Peptide on Human Colon Cancer (Caco-2) Cells. Molecules. 2022; 27(4):1426. https://doi.org/10.3390/molecules27041426
Chicago/Turabian StyleMaijaroen, Surachai, Sompong Klaynongsruang, Sittiruk Roytrakul, Monruedee Konkchaiyaphum, Lapatrada Taemaitree, and Nisachon Jangpromma. 2022. "An Integrated Proteomics and Bioinformatics Analysis of the Anticancer Properties of RT2 Antimicrobial Peptide on Human Colon Cancer (Caco-2) Cells" Molecules 27, no. 4: 1426. https://doi.org/10.3390/molecules27041426