Molecular Networking and Cultivation Profiling Reveals Diverse Natural Product Classes from an Australian Soil-Derived Fungus Aspergillus sp. CMB-MRF324
Abstract
:1. Introduction
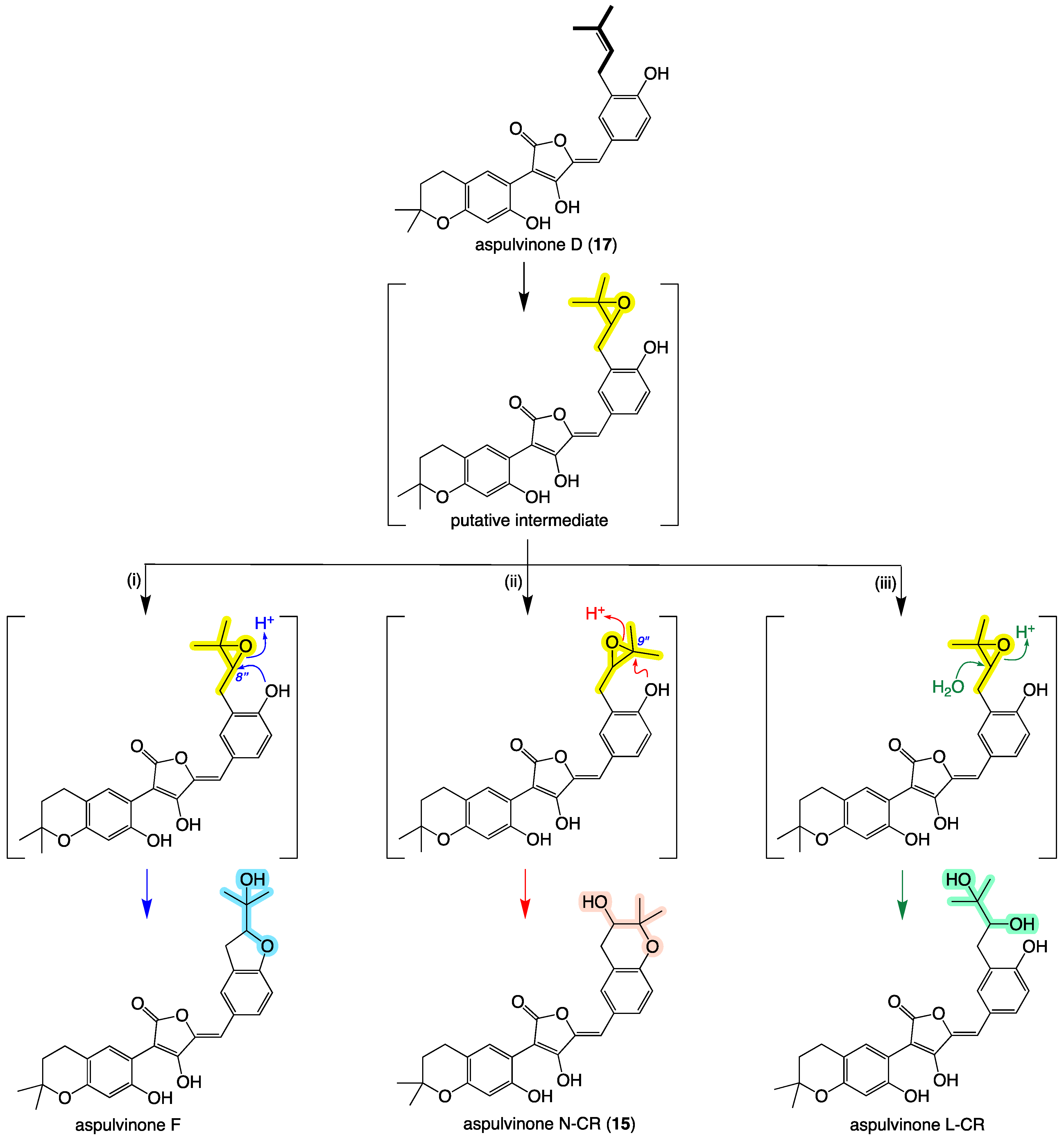
2. Results and Discussion
3. Materials and Methods
3.1. General Experimental Procedure
3.2. Collection and Taxonomy of CMB-MRF324
3.3. Scale-Up SDA Cultivation and Fractionation of CMB-MRF324
3.4. Scale-Up Brown Rice Cultivation and Fractionation of CMB-MRF324
3.5. Metabolite Characterization
3.6. Marfey’s Analysis of Aspergillamides E (7) and F (8)
3.7. Isomerisation of Aspergillamide F (8) to E (7)
3.8. X-ray Crystallography
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Capon, R.J.; Skene, C.; Stewart, M.; Ford, J.; O’Hair, R.A.J.; Williams, L.; Lacey, E.; Gill, J.H.; Heiland, K.; Friedel, T. Aspergillicins A–E: Five Novel Depsipeptides from the Marine-Derived Fungus Aspergillus carneus. Org. Biomol. Chem. 2003, 1, 1856–1862. [Google Scholar] [CrossRef] [PubMed]
- Capon, R.J.; Ratnayake, R.; Stewart, M.; Lacey, E.; Tennant, S.; Gill, J.H. Aspergillazines A-E: Novel Heterocyclic Dipeptides from an Australian Strain of Aspergillus unilateralis. Org. Biomol. Chem. 2005, 3, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Fremlin, L.J.; Piggott, A.M.; Lacey, E.; Capon, R.J. Cottoquinazoline A and Cotteslosins A and B, Metabolites from an Australian Marine-Derived Strain of Aspergillus versicolor. J. Nat. Prod. 2009, 72, 666–670. [Google Scholar] [CrossRef] [PubMed]
- Khalil, Z.G.; Huang, X.-C.; Raju, R.; Piggott, A.M.; Capon, R.J. Shornephine A: Structure, Chemical Stability, and P-Glycoprotein Inhibitory Properties of a Rare Diketomorpholine from an Australian Marine-Derived Aspergillus sp. J. Org. Chem. 2014, 79, 8700–8705. [Google Scholar] [CrossRef] [Green Version]
- Quezada, M.; Shang, Z.; Kalansuriya, P.; Salim, A.A.; Lacey, E.; Capon, R.J. Waspergillamide A, a Nitro Depsi-Tetrapeptide Diketopiperazine from an Australian Mud Dauber Wasp-Associated Aspergillus sp. (CMB-W031). J. Nat. Prod. 2017, 80, 1192–1195. [Google Scholar] [CrossRef]
- Aron, A.T.; Gentry, E.C.; McPhail, K.L.; Nothias, L.-F.; Nothias-Esposito, M.; Bouslimani, A.; Petras, D.; Gauglitz, J.M.; Sikora, N.; Vargas, F.; et al. Reproducible Molecular Networking of Untargeted Mass Spectrometry Data Using GNPS. Nat. Protoc. 2020, 15, 1954–1991. [Google Scholar] [CrossRef]
- Salim, A.A.; Khalil, Z.G.; Elbanna, A.H.; Wu, T.; Capon, R.J. Methods in Microbial Biodiscovery. Mar. Drugs 2021, 19, 503. [Google Scholar] [CrossRef]
- Wu, T.; Salim, A.A.; Capon, R.J. Millmerranones A-F: A Meroterpene Cyclic Carbonate and Related Metabolites from the Australian Fungus Aspergillus sp. CMB-MRF324. Org. Lett. 2021, 23, 8424–8428. [Google Scholar] [CrossRef]
- Alvi, K.A.; Pu, H.; Luche, M.; Rice, A.; App, H.; McMahon, G.; Dare, H.; Margolis, B. Asterriquinones Produced by Aspergillus candidus Inhibit Binding of the Grb-2 Adapter to Phosphorylated E Receptor Tyrosine Kinase. J. Antibiot. 1999, 52, 215–223. [Google Scholar] [CrossRef] [Green Version]
- Kaji, A.; Iwata, T.; Kiriyama, N.; Wakusawa, S.; Miyamoto, K.-I. Four New Metabolites of Aspergillus terreus. Chem. Pharm. Bull. 1994, 42, 1682. [Google Scholar] [CrossRef]
- Mocek, U.; Schultz, L.; Buchan, T.; Baek, C.; Fretto, L.; Nzerem, J.; Sehl, L.; Sinha, U. Isolation and Structure Elucidation of Five New Asterriquinones from Aspergillus, Humicola and Botryotrichum Species. J. Antibiot. 1996, 49, 854–859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakagawa, M.; Hirota, A.; Sakai, H.; Isogai, A. Terrecyclic Acid A, a New Antibiotic from Aspergillus terreus. I. Taxonomy, Production, and Chemical and Biological Properties. J. Antibiot. 1982, 35, 778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirota, A.; Nakagawa, M.; Sakai, H.; Isogai, A. Terrecyclic Acid A, a New Antibiotic from Aspergillus terreus. II. Structure of Terrecyclic Acid A. J. Antibiot. 1982, 35, 783. [Google Scholar] [CrossRef] [Green Version]
- Hirota, A.; Nakagawa, M.; Sakai, H.; Isogai, A. Terrecyclic Acid A, a New Antibiotic from Aspergillus terreus. III. Carbon-13 NMR Spectrum of Terrecyclic Acid A. J. Antibiot. 1984, 37, 475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirota, A.; Nakagawa, M.; Hirota, H.; Takahashi, T.; Isogai, A. Terrecyclic Acid A, a New Antibiotic from Aspergillus terreus. IV. Absolute Stereochemistry of Terrecyclic Acid A. J. Antibiot. 1986, 39, 149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cruz, P.G.; Auld, D.S.; Schultz, P.J.; Lovell, S.; Battaile, P.; MacArthur, R.; Shen, M.; Tamayo-Castillo, G.; Inglese, J.; Sherman, D.H. Titration-Based Screening for Evaluation of Natural Product Extracts: Identification of an Aspulvinone Family of Luciferase Inhibitors. Chem. Biol. 2011, 18, 1442–1452. [Google Scholar] [CrossRef] [Green Version]
- Ojima, N.; Takenaka, S.; Seto, S. New Butenolides from Aspergillus terreus. Phytochemistry 1973, 12, 2527–2529. [Google Scholar] [CrossRef]
- Ojima, N.; Takenaka, S.; Seto, S. Structures of Pulvinone Derivatives from Aspergillus terreus. Phytochemistry 1975, 14, 573–576. [Google Scholar] [CrossRef]
- Ojima, N.; Takahashi, I.; Ogura, K.; Seto, S. New Metabolites from Aspergillus terreus Related to the Biosynthesis of Aspulvinones. Tetrahedron Lett. 1976, 17, 1013–1014. [Google Scholar] [CrossRef]
- Capon, R.J. Extracting Value: Mechanistic Insights into the Formation of Natural Product Artifacts—Case Studies in Marine Natural Products. Nat. Prod. Rep. 2020, 37, 55–79. [Google Scholar] [CrossRef]
- Kagamizono, T.; Sakai, N.; Arai, K.; Kobinata, K.; Osada, H. Terpeptin, a Novel Mammalian Cell Cycle Inhibitor, Produced by Aspergillus terreus 95F-1. Tetrahedron Lett. 1997, 38, 1223–1226. [Google Scholar] [CrossRef]
- Toske, S.G.; Jensen, P.R.; Kauffman, C.A.; Fenical, W. Aspergillamides A and B: Modified Cytotoxic Tripeptides Produced by a Marine Fungus of the Genus Aspergillus. Tetrahedron 1998, 54, 13459–13466. [Google Scholar] [CrossRef]
- Shiomi, K.; Hatae, K.; Yamaguchi, Y.; Masuma, R.; Tomoda, H.; Kobayashi, S.; Omura, S. New Antibiotics Miyakamides Produced by a Fungus. J. Antibiot. 2002, 55, 952–961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Z.; Zhu, T.; Fang, Y.; Gu, Q. 1H and 13C NNMR Assignments of Two New Indolic Enamide Diastereomers from a Mangrove Endophytic Fungus Aspergillus sp. Magn. Reson. Chem. 2008, 46, 1212–1216. [Google Scholar] [CrossRef]
- Izumikawa, M.; Hashimoto, J.; Takagi, M.; Shin-ya, K. Isolation of Two New Terpeptin Analogs—JBIR-81 and JBIR-82—From a Seaweed-Derived Fungus, Aspergillus sp. SpD081030G1f1. J. Antibiot. 2010, 63, 389–391. [Google Scholar] [CrossRef] [Green Version]
- Luo, X.W.; Lin, Y.; Lu, Y.-J.; Zhou, X.-F.; Liu, Y.-H. Peptides and Polyketides Isolated from the Marine Sponge-Derived Fungus Aspergillus terreus SCSIO 41008. Chin. J. Nat. Med. 2019, 17, 149–154. [Google Scholar] [CrossRef]
- Hooft, R.W.W.; Straver, L.H.; Spek, A.L. Determination of Absolute Structure Using Bayesian Statistics on Bijvoet Differences. J. Appl. Crystallogr. 2008, 41, 96–103. [Google Scholar] [CrossRef] [Green Version]
- Neff, S.A.; Lee, S.U.; Asami, Y.; Ahn, J.S.; Oh, H.; Baltrusaitis, J.; Gloer, J.B.; Wicklow, D.T. Aflaquinolones A-G: Secondary Metabolites from Marine and Fungicolous Isolates of Aspergillus spp. J. Nat. Prod. 2012, 75, 464–472. [Google Scholar] [CrossRef] [Green Version]
- Mou, X.-F.; Liu, X.; Xu, R.-F.; Wei, M.-Y.; Fang, Y.W.; Shao, C.-L. Scopuquinolone B, a New Monoterpenoid Dihydroquinolin-2(1H)-One Isolated from the Coral-Derived Scopulariopsis sp. Fungus. Nat. Prod. Rep. 2018, 32, 773–776. [Google Scholar] [CrossRef]
- Chen, M.; Shao, C.-L.; Meng, H.; She, Z.-G.; Wang, C.-Y. Anti-Respiratory Syncytial Virus Prenylated Dihydroquinolone Derivatives from the Gorgonian-Derived Fungus Aspergillus sp. XS-20090B15. J. Nat. Prod. 2014, 77, 2720–2724. [Google Scholar] [CrossRef]
- Wu, C.; Cui, X.; Sun, L.; Lu, J.; Li, F.; Song, M.; Zhang, Y.; Hao, X.; Tian, C.; Song, M.; et al. Aspulvinones Suppress Postprandial Hyperglycemia as Potent A-Glucosidase Inhibitors from Aspergillus terreus ASM-1. Front. Chem. 2021, 9, 736070. [Google Scholar] [CrossRef] [PubMed]
- Elyashberg, M.; Blinov, K.; Molodtsov, S.; Williams, A.J. Structure Revision of Asperjinone Using Computer-Assisted Structure Elucidation Methods. J. Nat. Prod. 2013, 76, 113–116. [Google Scholar] [CrossRef]
- Begley, M.J.; Gedge, D.R.; Knight, D.W.; Pattenden, G. Aspulvinones, a New Class of Natural Products from Aspergillus terreus. Re-Investigation of Structures by X-Ray Crystallographic and Spectroscopic Analysis. J. Chem. Soc., Perkin Trans. 1979, 1, 77–83. [Google Scholar] [CrossRef]
- Gao, H.; Guo, W.; Wang, Q.; Zhang, L.; Zhu, M.; Zhu, T.; Gu, Q.; Wang, W.; Li, D. Aspulvinones from a Mangrove Rhizosphere Soil-Derived Fungus Aspergillus terreus Gwq-48 with Anti-Influenza A Viral (H1N1) Activity. Bioorg. Med. Chem. Lett. 2013, 23, 1776–1778. [Google Scholar] [CrossRef]
- Guo, C.-J.; Knox, B.P.; Sanchez, J.F.; Chiang, Y.-M.; Bruno, K.S.; Wang, C.C.C. Application of an Efficient Gene Targeting System Linking Secondary Metabolites to Their Biosynthetic Genes in Aspergillus terreus. Org. Lett. 2013, 15, 3562–3565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, P.; Li, X.-M.; Wang, J.-N.; Li, X.; Wang, B.-G. New Butenolide Derivatives from the Marine-Derived Fungus Paecilomyces variotii with DPPH Radical Scavenging Activity. Phytochem. Lett. 2015, 11, 85–88. [Google Scholar] [CrossRef]
- Zhang, L.-H.; Feng, B.-M.; Zhao, Y.-Q.; Sun, Y.; Liu, B.; Liu, F.; Chen, G.; Bai, J.; Hua, H.-M.; Wang, H.-F.; et al. Polyketide Butenolide, Diphenyl Ether, and Benzophenone Derivatives from the Fungus Aspergillus flavipes PJ03-11. Bioorg. Med. Chem. Lett. 2016, 26, 346–350. [Google Scholar] [CrossRef]
- Pang, X.; Zhao, J.-Y.; Fang, X.-M.; Zhang, T.; Zhang, D.-W.; Liu, H.-Y.; Su, J.; Cen, S.; Yu, L.-Y. Metabolites from the Plant Endophytic Fungus Aspergillus sp. CPCC 400735 and Their Anti-HIV Activities. J. Nat. Prod. 2017, 80, 2595–2601. [Google Scholar] [CrossRef]
- Machado, F.P.; Kumla, D.; Pereira, J.A.; Sousa, E.; Dethoup, T.; Freitas-Silva, J.; Costa, P.M.; Mistry, S.; Silva, A.M.S.; Kijjoa, A. Prenylated Phenylbutyrolactones from Cultures of a Marine Sponge-Associated Fungus Aspergillus flavipes KUFA1152. Phytochemistry 2021, 185, 112709. [Google Scholar] [CrossRef]
- Liang, X.-X.; Zhang, X.-J.; Zhao, Y.-X.; Feng, J.; Zeng, J.-C.; Shi, Q.-Q.; Kaunda, J.S.; Li, X.-L.; Wang, W.-G.; Xiao, W.-L. Aspulvins A–H, Aspulvinone Analogues with SARS-CoV-2 Mpro Inhibitory and Anti-Inflammatory Activities from an Endophytic Cladosporium sp. J. Nat. Prod. 2022, 85, 878–887. [Google Scholar] [CrossRef]
- Hsiao, G.; Chi, W.-C.; Chang, C.-H.; Chiang, Y.-R.; Fu, Y.-J.; Lee, T.-H. Bioactive Pulvinones from a Marine Algicolous Fungus Aspergillus Terreus NTU243. Phytochemistry 2022, 200, 113229. [Google Scholar] [CrossRef] [PubMed]
- Manchoju, A.; Annadate, R.A.; Desquien, L.; Pansare, S.V. Functionalization of Diazotetronic Acid and Application in a Stereoselective Modular Synthesis of Pulvinone, Aspulvinones A–E, G, Q and Their Analogues. Org. Biomol. Chem. 2018, 16, 6224–6238. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.-Y.; Ruan, B.-H.; Yang, Y.-B.; Wang, Q.; Li, X.-Z.; Luo, N.; Yang, X.-Q.; Zhao, L.-X. Secondary Metabolites of the Fungus Aspergillus ferreus. Chem. Nat. Compd. 2018, 54, 415–418. [Google Scholar] [CrossRef]
- Sheldrick, G.M. A Short History of Shelx. Acta Crystallogr., Sect. A Found. Crystallogr. 2008, 64, 112–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macrae, C.F.; Edgington, P.R.; McCabe, P.; Pidcock, E.; Shields, G.P.; Taylor, R.; Towler, M.; van de Streek, J. Mercury: Visualization and Analysis of Crystal Structures. J. Appl. Crystallogr. 2006, 39, 453–457. [Google Scholar] [CrossRef] [Green Version]
- Farrugia, L.J. Wingx Suite for Small-Molecule Single-Crystal Crystallography. J. Appl. Crystallogr. 1999, 32, 837–838. [Google Scholar] [CrossRef]

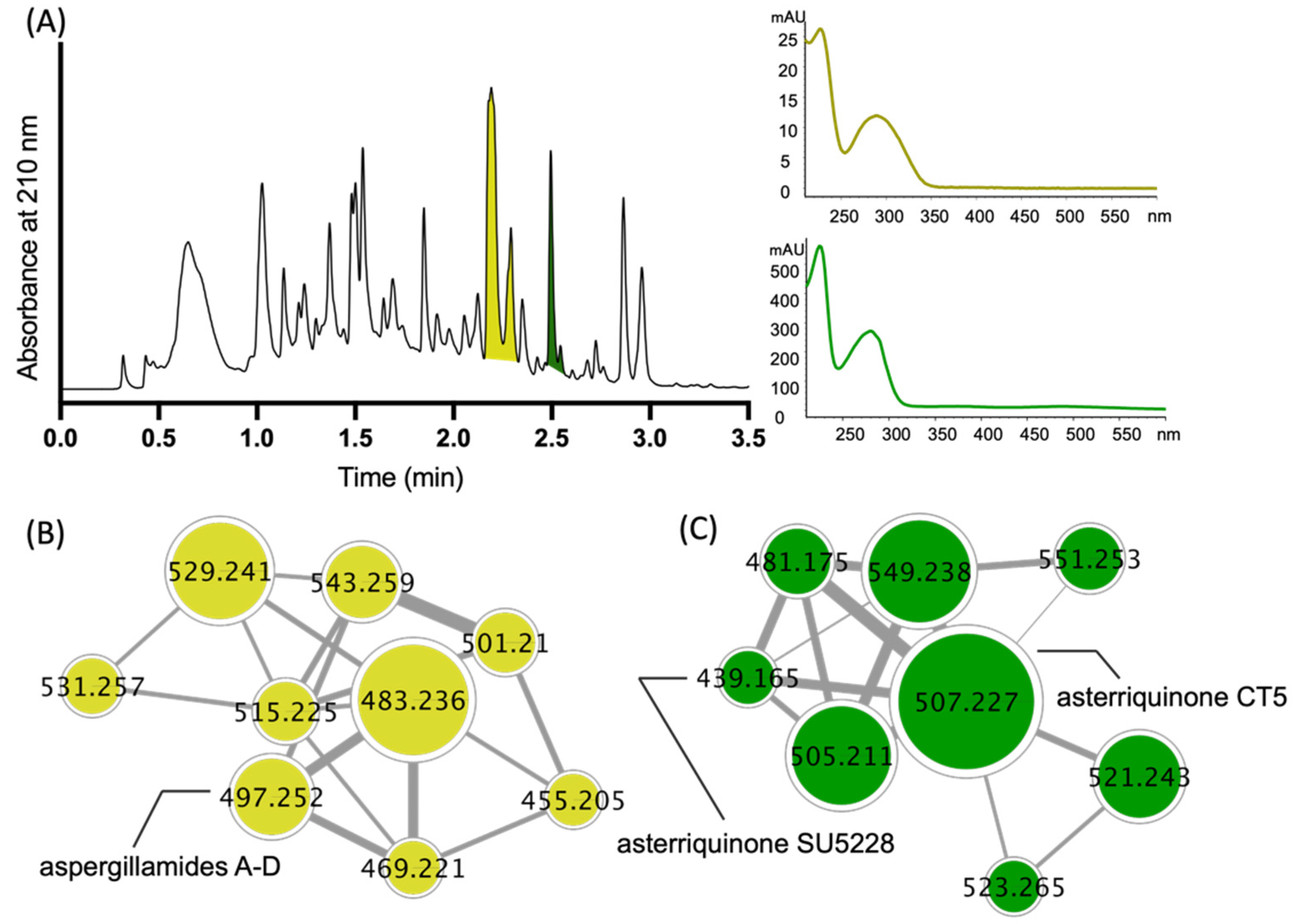
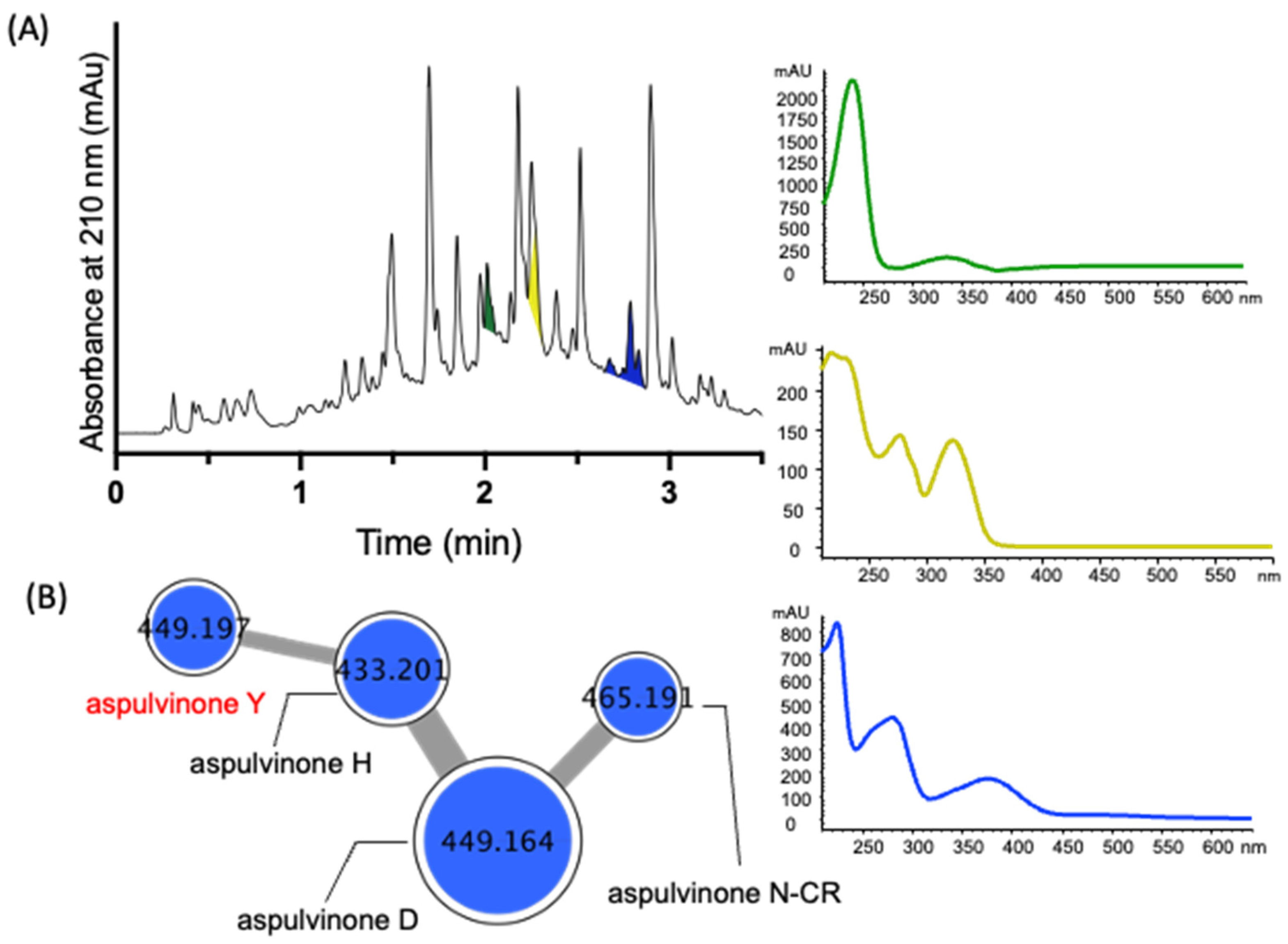
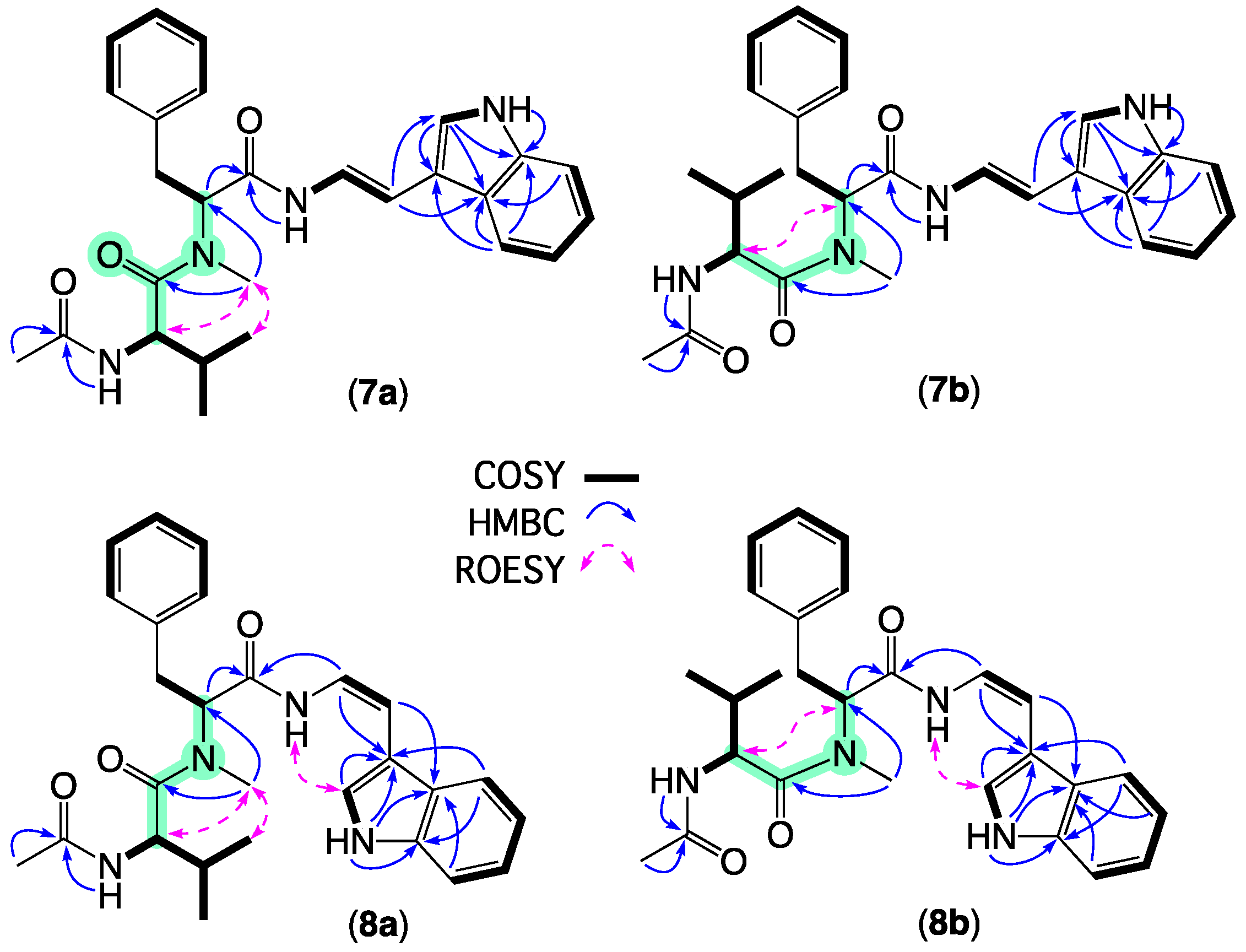
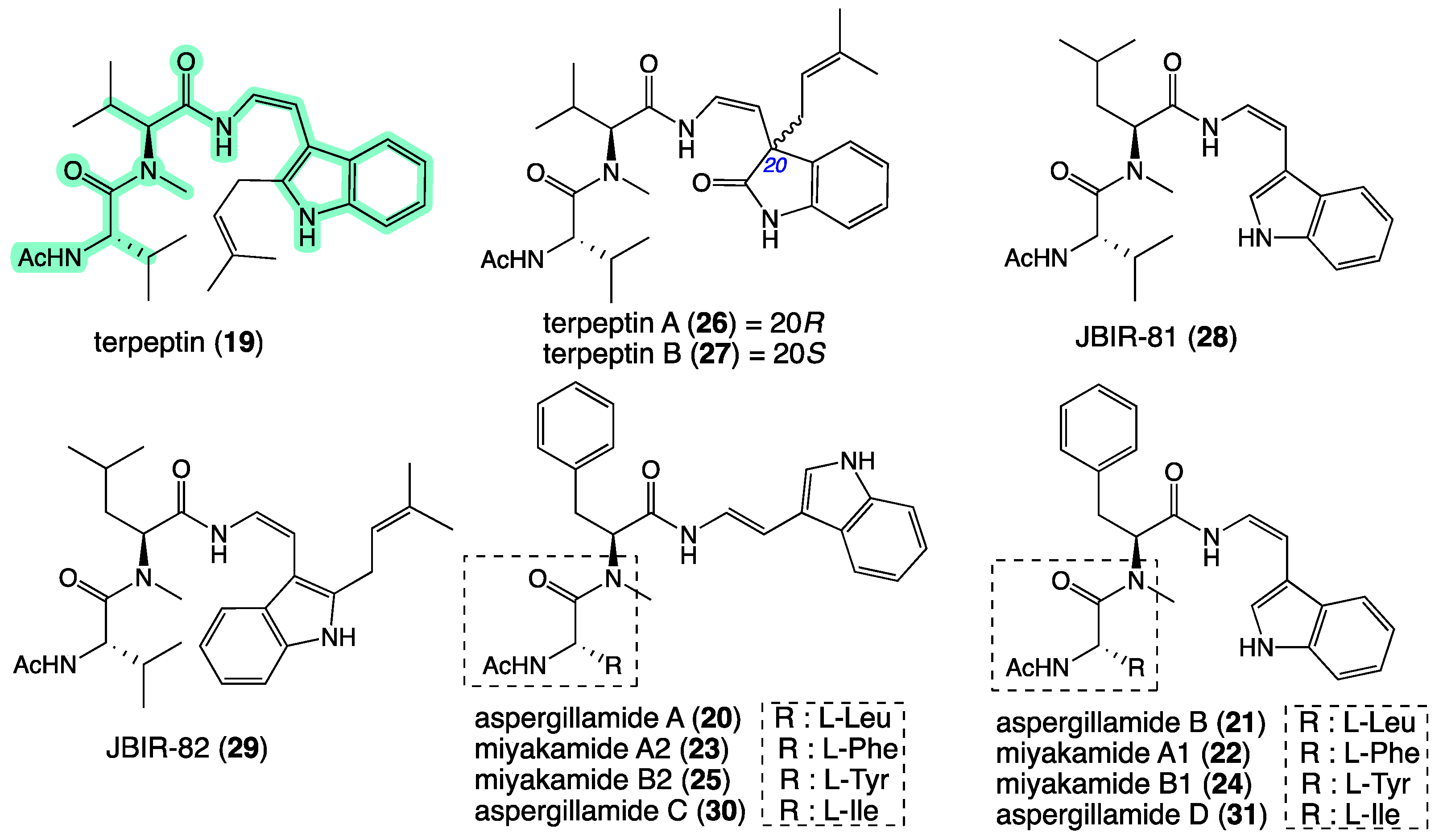
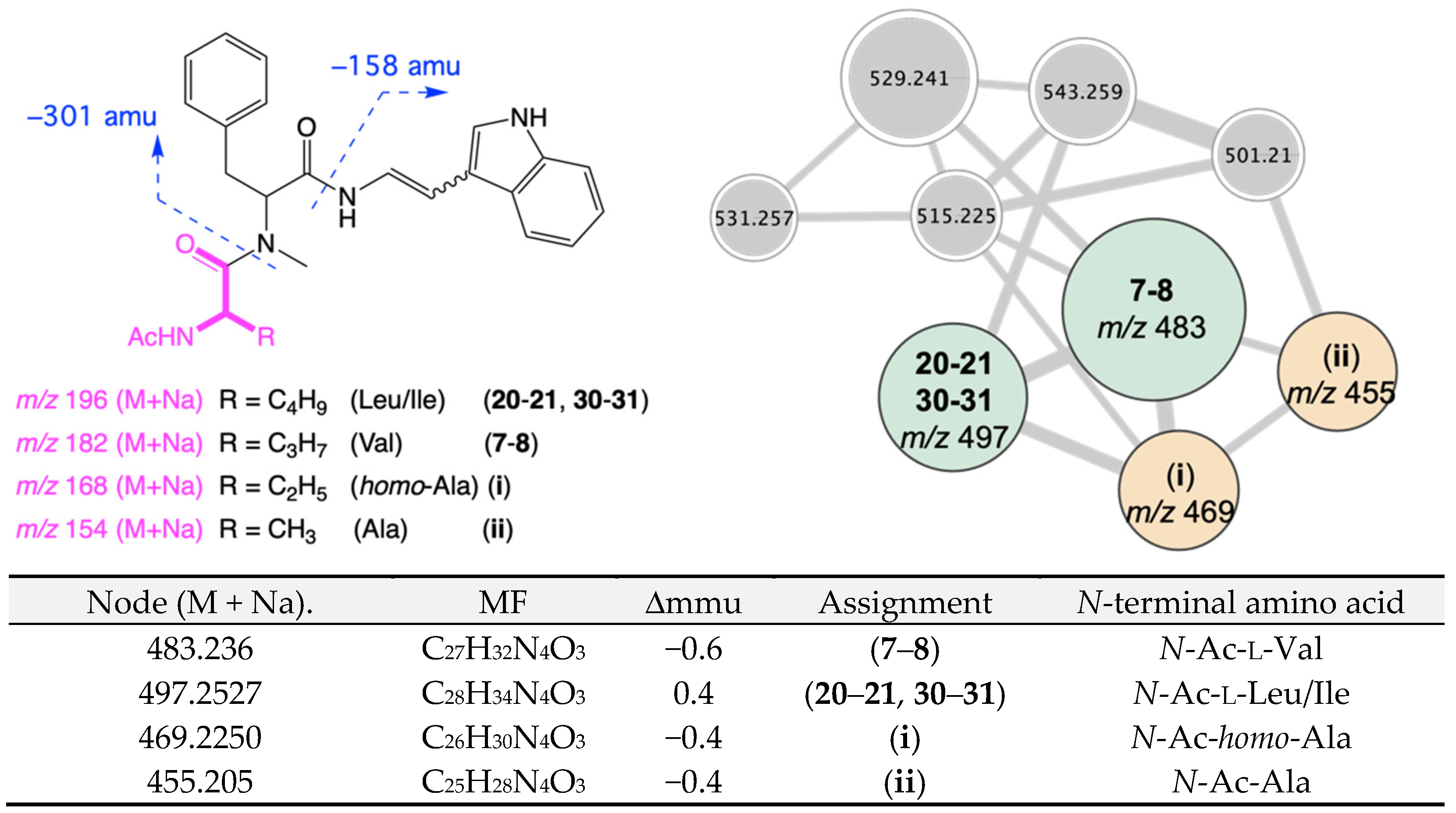
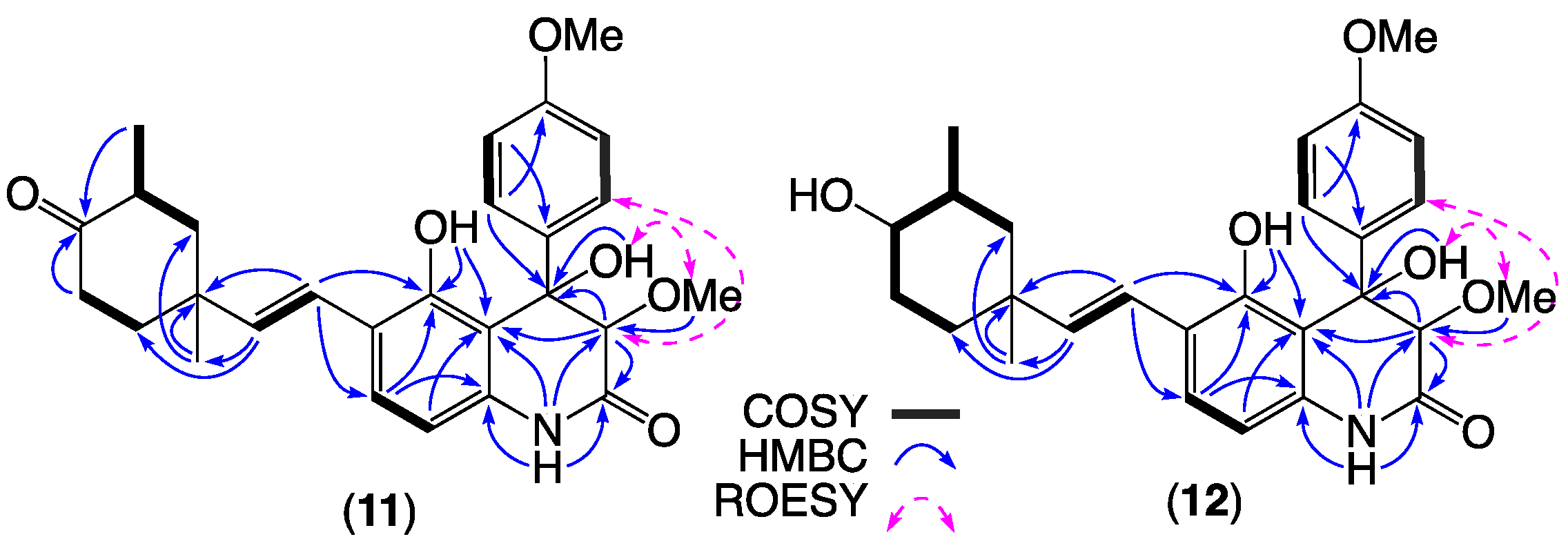

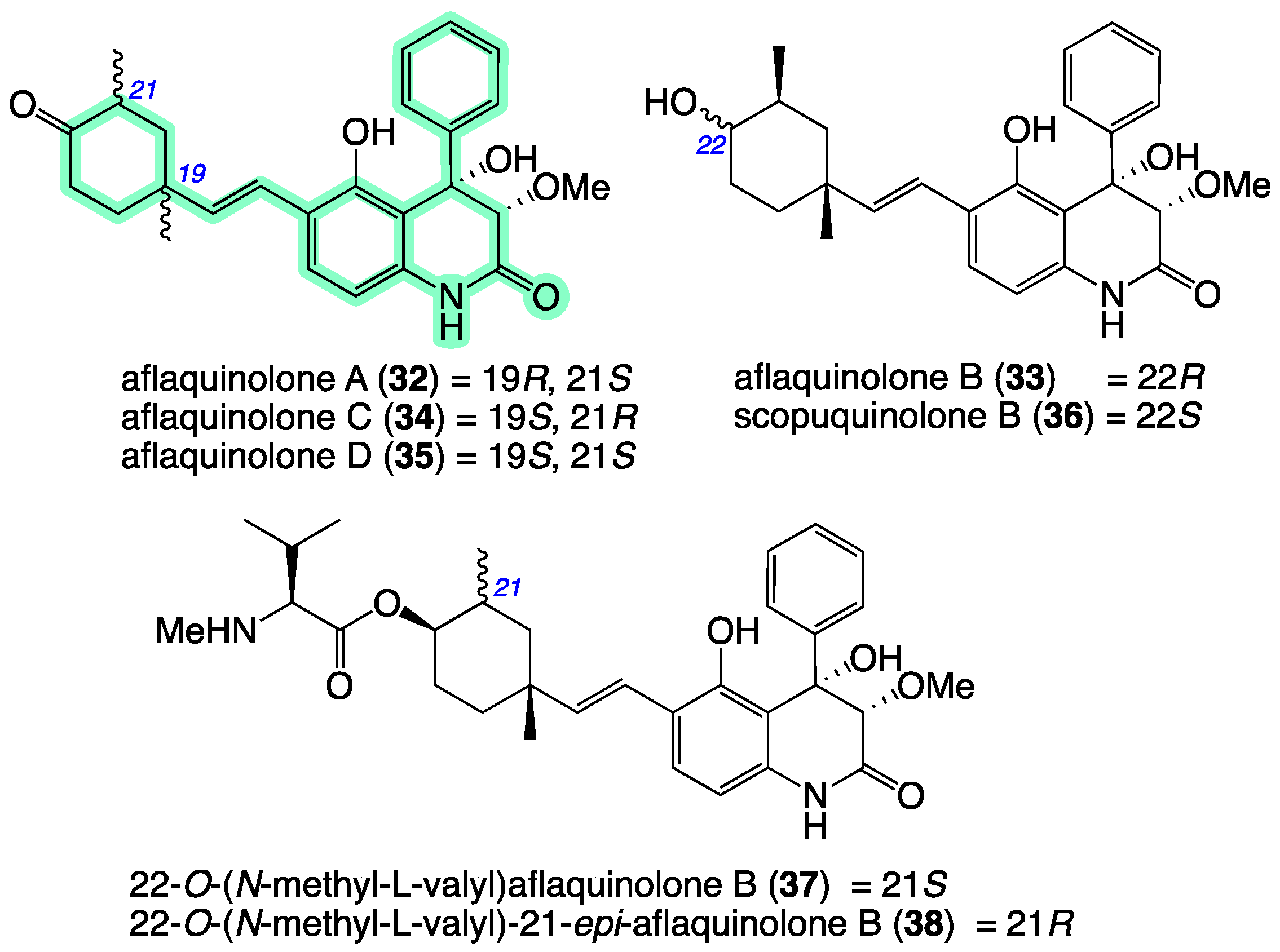

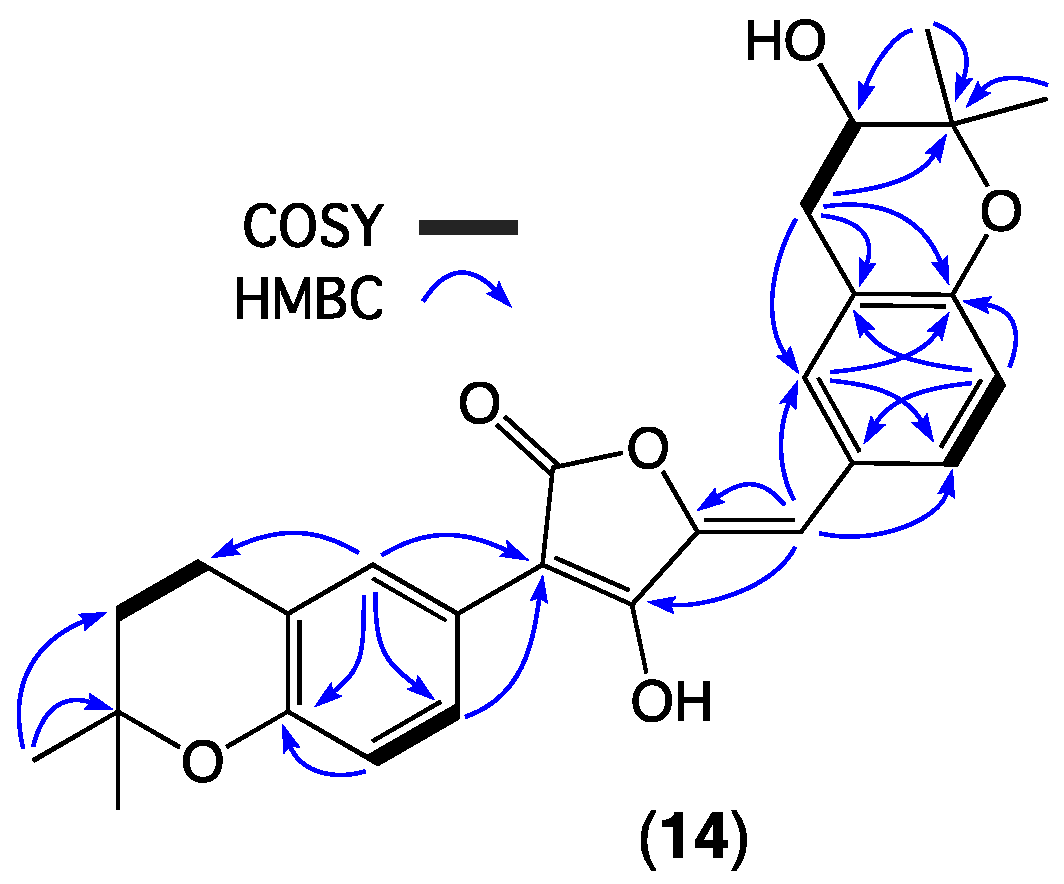
| Position | 7a δH, Mult (J in Hz) | 7b δH, Mult (J in Hz) | 8a δH, Mult (J in Hz) | 8b δH, Mult (J in Hz) |
|---|---|---|---|---|
| enamino-Trp | ||||
| 2 | 7.42, d (2.2) | 7.45, d (2.2) | 7.53, d (2.3) | 7.71, d (2.3) |
| 4 | 7.61, d (7.7) | 7.62, d (7.7) | 7.57, d (7.9) | 7.57 B |
| 5 | 7.07, dd (7.7, 7.7) | 7.11, m | 7.02, dd (7.9, 7.3) | 7.03 B |
| 6 | 7.12, m | 7.07, m | 7.12, dd (7.9, 7.3) | 7.12 B |
| 7 | 7.38, d (8.0) | 7.36, d (8.0) | 7.38, d (7.9) | 7.37, d (8.3) |
| 8 | 6.42, d (15.0) | 6.52, d (15.0) | 5.95, d (9.5) | 5.99, d (9.5) |
| 9 | 7.27, dd (15.0, 9.9) | 7.32, dd (15.0, 9.9) | 6.64, dd (10.2, 9.5) | 6.61, dd (9.8, 9.5) |
| 1-NH | 11.1, d (2.2) | 11.1, d (2.2) | 11.29, br s | 11.30, br s |
| 9-NH | 9.93, d (9.9) | 9.92, d (9.9) | 9.01, d (10.2) | 9.23, d (9.8) |
| N-Me-l-Phe | ||||
| 2 | 5.27, dd (9.6, 6.1) | 4.98, dd (9.6, 5.2) | 5.53, dd (9.9, 6.0) | 5.30, dd (8.8, 6.0) |
| 3 | a. 3.23, dd (14.3, 6.1) | a. 3.32 A | a. 3.23, dd (15.0, 6.0) | a. 3.37 A |
| b. 2.97, dd (14.3, 9.6) | b. 2.95, d (14.3, 6.1) | b. 3.00, dd (15.0, 9.9) | b. 2.96, dd (14.4, 8.8) | |
| 5 | 7.21, d (7.1) | 7.29, d (7.1) | 7.23, d (7.1) | 7.30, d (7.0) |
| 6 | 7.24, dd (7.1, 7.1) | 7.30, dd (7.1, 7. 1) | 7.22, dd (7.1, 7.1) | 7.29, dd (7.0, 7.0) |
| 7 | 7.18, t (7.1) | 7.22, t (7.1) | 7.17, t (7.1) | 7.22 B |
| N-Me | 3.08, s | 2.81, s | 3.03, s | 2.89, s |
| l-Val | ||||
| 2 | 4.44, dd (8.7, 8.7) | 4.29, dd (7.6, 7.6) | 4.46, dd (8.9, 8.9) | 4.26, dd (8.1, 8.1) |
| 3 | 1.90, dqq (8.7, 6.9, 6.9) | 1.23, dqq (7.6, 6.9, 6.9) | 1.89, dqq (8.9, 6.7, 6.7) | 1.56, dqq (8.1, 6.8, 6.8) |
| 4C | 0.81, d (6.9) | 0.63, d (6.9) | 0.72, d (6.7) | 0.48, d (6.8) |
| 5C | 0.84, d (6.9) | 0.48, d (6.9) | 0.77, d (6.7) | 0.69, d (6.8) |
| NH | 7.97, d (8.7) | 8.26, d (7.6) | 7.9, d (8.9) | 8.24, d (8.1) |
| NHAc | 1.79, s | 1.91, s | 1.73, s | 1.66, s |
| Position | 7a δC, Type | 7b δC, Type | 8a δC, Type | 8b δC, Type |
|---|---|---|---|---|
| enamino-Trp | ||||
| 2 | 123.5, CH | 123.6, CH | 123.6, CH | 124.4, CH |
| 3 | 111.5, C | 111.4, C | 109.5, C | 109.2, C |
| 3a | 124.8, C | 124.9, C | 126.5, C | 126.6, C |
| 4 | 118.9, CH | 118.8, CH | 118.3, CH | 118.3, CH |
| 5 | 119.3, CH | 119.3, CH | 119.1, CH | 119.1, CH |
| 6 | 121.4, CH | 121.4, CH | 121.6, CH | 121.5, CH |
| 7 | 111.8, CH | 111.8, CH | 111.5, CH | 111.4, CH |
| 7a | 136.8, C | 136.8, C | 135.6, C | 135.6, C |
| 8 | 106.8, CH | 107.0, CH | 103.3, CH | 104.2, CH |
| 9 | 119.5, CH | 119.5, CH | 118.1, CH | 117.9, CH |
| N-Me-l-Phe | ||||
| 1 | 167.0, C | 166.1, C | 168.4, C | 167.6, C |
| 2 | 57.7, CH | 61.3, CH | 56.8, CH | 60.6, CH |
| 3 | 34.1, CH2 | 34.2, CH2 | 33.5, CH2 | 35.4, CH2 |
| 4 | 137.6, C | 137.8, C | 137.4, C | 137.5, C |
| 5 | 128.8, CH | 129.3, CH | 128.8, CH | 129.3, CH |
| 6 | 128.1, CH | 128.4, CH | 128.1, CH | 128.3, CH |
| 7 | 126.3, CH | 126.5, CH | 126.3, CH | 126.5, CH |
| N-Me | 32.3, CH3 | 29.3, CH3 | 31.6, CH3 | 29.7, CH3 |
| l-Val | ||||
| 1 | 172.4, C | 171.5, C | 172.8, C | 171.8, C |
| 2 | 54.0, CH | 53.6, CH | 53.6, CH | 54.0, CH |
| 3 | 30.0, CH | 28.7, CH | 30.0, CH | 29.2, CH |
| 4 A | 18.4, CH3 | 17.5, CH3 | 18.9, CH3 | 19.3, CH3 |
| 5 A | 18.9, CH3 | 19.3, CH3 | 18.2, CH3 | 18.0, CH3 |
| NHCOCH3 | 169.1, C | 170.6, C | 168.8, C | 170.2, C |
| NHCOCH3 | 22.2, CH3 | 22.2, CH3 | 22.1, CH3 | 21.7, CH3 |
| Position | 11 | 12 | ||
|---|---|---|---|---|
| δH, Mult (J in Hz) | δC, Type | δH, Mult (J in Hz) | δC, Type | |
| 2 | - | 165.8, C | - | 165.4, C |
| 3 | 3.70, br d (1.5) | 84.3, CH | 3.69, br d (1.4) | 84.4, CH |
| 3-OMe | 3.61, s | 59.1, C | 3.60, s | 59.0, C |
| 4 | - | 78.9, C | - | 78.9, C |
| 5 | - | 111.1, C | - | 111.0, C |
| 6 | - | 155.2, C | - | 155.0, C |
| 7 | - | 122.4, C | - | 123.0, C |
| 8 | 7.40, d (8.2) | 127.3, CH | 7.37, d (8.1) | 127.1, CH |
| 9 | 6.36, d (8.2) | 107.0, CH | 6.32, d (8.1) | 106.8, CH |
| 10 | - | 134.4, C | - | 134.0, C |
| 11 | - | 129.2, C | - | 129.2, C |
| 12 | 7.18, d (8.8) | 128.0, CH | 7.18, d (8.8) | 128.0, CH |
| 13 | 6.82, d (8.8) | 114.5, CH | 6.83, d (8.8) | 114.5, CH |
| 14 | - | 160.5, C | - | 160.4, C |
| 14-OMe | 3.76, s | 55.4, CH3 | 3.76, s | 55.5, CH3 |
| 15 | 6.82, d (8.8) | 114.5, CH | 6.83, d (8.8) | 114.5, CH |
| 16 | 7.18, d (8.8) | 128.0, CH | 7.18, d (8.8) | 128.0, CH |
| 17 | 6.79, d (16.6) | 122.7, CH | 6.61, d (16.8) | 121.8, CH |
| 18 | 6.28, d (16.6) | 135.9, CH | 6.14, d (16.8) | 137.7, CH |
| 19 | - | 37.6, C | - | 37.2, C |
| 20 | a. 2.11, m | 47.7, CH2 | a. 1.74, m | 46.1, CH2 |
| b. 1.47, dd (13.4, 13.4) | b. 1.07, m | |||
| 21 | 2.54, m | 41.5, CH | 1.57 A | 36.3, CH |
| 22 | - | 214.0, C | 3.12, ddd (10.5, 10.5, 4.5) | 77.3, CH |
| 23 | a. 2.49, dd (14.2, 5.7) | 38.7, CH2 | a. 1.77, m | 32.1, CH2 |
| b. 2.24, m | b. 1.51 A | |||
| 24 | a. 2.15, m | 38.5, CH2 | a. 1.82, m | 36.9, CH2 |
| b. 1.73, td (13.4, 4.4) | b. 1.37, td (13.7, 3.5) | |||
| 25 | 1.10, s | 30.6, CH3 | 1.03, s | 31.3, CH3 |
| 26 | 0.99, d (6.5) | 14.6, CH3 | 0.98, d (6.4) | 18.8, CH3 |
| 1-NH | 7.75, br s | - | 7.35, br s | - |
| 4-OH | 4.58, s | - | 4.54, s | - |
| 6-OH | 9.14, s | - | 9.08, s | - |
| Position | 14 | |
|---|---|---|
| δH, Mult (J in Hz) | δC, Type | |
| 1 | - | ND |
| 2 | - | 101.6 |
| 3 | - | 164.2 |
| 4 | - | 142.3 |
| 5 | 6.38, s | 108.0 |
| 1′ | - | 122.6 |
| 2′ | 7.64, d (1.5) | 129.7 |
| 3′ | - | 121.7 |
| 4′ | - | 154.3 |
| 5′ | 6.73, d (8.3) | 117.6 |
| 6′ | 7.62, dd (8.3, 1.5) | 127.7 |
| 7′ | 2.83, t (6.8) | 23.1 |
| 8′ | 1.84, t (6.8) | 33.5 |
| 9′ | - | 75.2 |
| 10′ | 1.33, s | 26.8 |
| 11′ | 1.33, s | 26.8 |
| 1″ | - | 126.3 |
| 2″ | 7.57, s | 133.1 |
| 3″ | - | 121.4 |
| 4″ | - | 155.0 |
| 5″ | 6.78, d (8.6) | 118.1 |
| 6″ | 7.51, d (1.5) | 130.9 |
| 7″ | a. 3.07, dd (16.4, 5.0) | 31.8 |
| b. 2.78, dd (16.4, 7.3) | ||
| 8″ | 3.79, dd (7.3, 5.0) | 69.9 |
| 9″ | - | 78.5 |
| 10″ | 1.29, s | 21.1 |
| 11″ | 1.35, s | 25.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, T.; Salim, A.A.; Bernhardt, P.V.; Capon, R.J. Molecular Networking and Cultivation Profiling Reveals Diverse Natural Product Classes from an Australian Soil-Derived Fungus Aspergillus sp. CMB-MRF324. Molecules 2022, 27, 9066. https://doi.org/10.3390/molecules27249066
Wu T, Salim AA, Bernhardt PV, Capon RJ. Molecular Networking and Cultivation Profiling Reveals Diverse Natural Product Classes from an Australian Soil-Derived Fungus Aspergillus sp. CMB-MRF324. Molecules. 2022; 27(24):9066. https://doi.org/10.3390/molecules27249066
Chicago/Turabian StyleWu, Taizong, Angela A. Salim, Paul V. Bernhardt, and Robert J. Capon. 2022. "Molecular Networking and Cultivation Profiling Reveals Diverse Natural Product Classes from an Australian Soil-Derived Fungus Aspergillus sp. CMB-MRF324" Molecules 27, no. 24: 9066. https://doi.org/10.3390/molecules27249066
APA StyleWu, T., Salim, A. A., Bernhardt, P. V., & Capon, R. J. (2022). Molecular Networking and Cultivation Profiling Reveals Diverse Natural Product Classes from an Australian Soil-Derived Fungus Aspergillus sp. CMB-MRF324. Molecules, 27(24), 9066. https://doi.org/10.3390/molecules27249066







