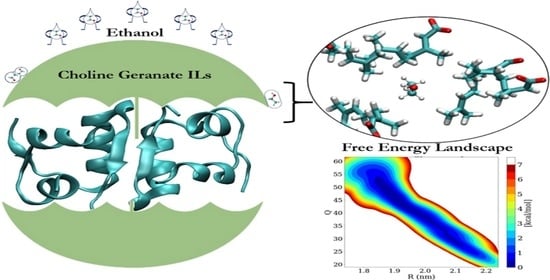
Counteractive Effects of Choline Geranate (CAGE) ILs and Ethanol on Insulin’s Stability—A Leap Forward towards Oral Insulin Formulation
Abstract
:1. Introduction
2. Computational Details and Methods
2.1. Molecular Dynamics (MD) Simulations
2.2. Well-Tempered Metadynamics (WT-MetaD) Simulations
3. Results and Discussion
3.1. Structural Stability of Insulin Dimer with CAGE and Ethanol
3.2. Accumulation of CAGE, Ethanol, and Water Molecules around the Insulin Dimer
3.3. Interactions of Choline, Geranate, and Ethanol with Water Molecules
3.4. Interactions between CAGE and Ethanol
3.5. Binding Free Energy (∆Gbinding) Calculations
3.6. Free Energy Landscape (FEL) of Insulin Dimer
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schreiber, G.; Haran, G.; Zhou, H.X. Fundamental Aspects of Protein—Protein Association Kinetics. Chem. Rev. 2009, 109, 839–860. [Google Scholar] [CrossRef] [Green Version]
- Banerjee, P.; Bagchi, B. Dynamical Control by Water at a Molecular Level in Protein Dimer Association and Dissociation. Proc. Natl. Acad. Sci. USA 2020, 117, 2302–2308. [Google Scholar] [CrossRef]
- Marianayagam, N.J.; Sunde, M.; Matthews, J.M. The Power of Two: Protein Dimerization in Biology. Trends Biochem. Sci. 2004, 29, 618–625. [Google Scholar] [CrossRef]
- Urbanc, B. Protein Actions: Principles and Modeling by I. Bahar, R.L. Jernigan, and K.A. Dill. J. Biol. Phys. 2017, 43, 585–589. [Google Scholar] [CrossRef]
- Barducci, A.; Bonomi, M.; Prakash, M.K.; Parrinello, M. Free-Energy Landscape of Protein Oligomerization from Atomistic Simulations. Proc. Natl. Acad. Sci. USA 2013, 110, E4708–E4713. [Google Scholar] [CrossRef] [Green Version]
- Brogden, R.N.; Heel, R.C. Human Insulin: A Review of Its Biological Activity, Pharmacokinetics and Therapeutic Use. Drugs 1987, 34, 350–371. [Google Scholar] [CrossRef]
- Roglic, G. WHO Global Report on Diabetes: A Summary. Int. J. Noncommun. Dis. 2016, 1, 3. [Google Scholar] [CrossRef]
- Knegtel, R.M.A.; Boelens, R.; Ganadu, M.L.; Kaptein, R. The Solution Structure of Monomeric Insulin. Eur. J. Biochem. 1991, 202, 447–458. [Google Scholar] [CrossRef]
- Brange, J.; Langkjoer, L. Insulin Structure and Stability. Pharm. Biotechnol. 1993, 5, 315–350. [Google Scholar] [CrossRef]
- Gualandi-Signorini, A.M.; Giorgi, G. Insulin Formulations—A Review. Eur. Rev. Med. Pharmacol. Sci. 2001, 5, 73–83. [Google Scholar]
- Galloway, J.A.; Chance, R.E. Improving Insulin Therapy: Achievements and Challenges. Horm. Metab. Res. 1994, 26, 591–598. [Google Scholar] [CrossRef]
- Pillai, O.; Panchagnula, R. Insulin Therapies—Past, Present and Future. Drug Discov. Today 2001, 6, 1056–1061. [Google Scholar] [CrossRef]
- Fu, A.Z.; Qiu, Y.; Radican, L. Impact of Fear of Insulin or Fear of Injection on Treatment Outcomes of Patients with Diabetes. Curr. Med. Res. Opin. 2009, 25, 1413–1420. [Google Scholar] [CrossRef]
- Arbit, E.; Kidron, M. Oral Insulin: The Rationale for This Approach and Current Developments. J. Diabetes Sci. Technol. 2009, 3, 562–567. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Yu, J.; Kahkoska, A.R.; Wang, J.; Buse, J.B.; Gu, Z. Advances in Transdermal Insulin Delivery. Adv. Drug Deliv. Rev. 2019, 139, 51–70. [Google Scholar] [CrossRef]
- Fonte, P.; Araújo, F.; Reis, S.; Sarmento, B. Oral Insulin Delivery: How Far Are We? J. Diabetes Sci. Technol. 2013, 7, 520–531. [Google Scholar] [CrossRef] [Green Version]
- Sonia, T.A.; Sharma, C.P. Oral Delivery of Insulin. Oral Deliv. Insul. 2015, 6, 1–345. [Google Scholar] [CrossRef]
- Palanisamy, K.; Prakash, M.; Rajapandian, V. Combined DFT and MD Simulation Studies of Protein Stability on Imidazolium–Water (ImH+Wn) Clusters with Aromatic Amino Acids. New J. Chem. 2020, 44, 17912–17923. [Google Scholar] [CrossRef]
- Sivapragasam, M.; Moniruzzaman, M.; Goto, M. Recent Advances in Exploiting Ionic Liquids for Biomolecules: Solubility, Stability and Applications. Biotechnol. J. 2016, 11, 1000–1013. [Google Scholar] [CrossRef]
- Egorova, K.S.; Gordeev, E.G.; Ananikov, V.P. Biological Activity of Ionic Liquids and Their Application in Pharmaceutics and Medicine. Chem. Rev. 2017, 117, 7132–7189. [Google Scholar] [CrossRef]
- Petkovic, M.; Ferguson, J.L.; Gunaratne, H.Q.N.; Ferreira, R.; Leitão, M.C.; Seddon, K.R.; Rebelo, L.P.N.; Pereira, C.S. Novel Biocompatible Cholinium-Based Ionic Liquids—Toxicity and Biodegradability. Green Chem. 2010, 12, 643–664. [Google Scholar] [CrossRef]
- Santos, J.I.; Gonçalves, A.M.M.; Pereira, J.L.; Figueiredo, B.F.H.T.; Silva, F.A.E.; Coutinho, J.A.P.; Ventura, S.P.M.; Gonçalves, F. Environmental Safety of Cholinium-Based Ionic Liquids: Assessing Structure-Ecotoxicity Relationships. Green Chem. 2015, 17, 4657–4668. [Google Scholar] [CrossRef]
- Palanisamy, K.; Rubavathy, S.M.E.; Prakash, M.; Thilagavathi, R.; Hosseini-zare, M.S.; Selvam, C. RSC Advances Antiviral Activities of Natural Compounds and Ionic Liquids to Inhibit the Mpro of SARS-CoV-2: A Computational Approach. RSC Adv. 2022, 12, 3687–3695. [Google Scholar] [CrossRef]
- Sundaram, V.; Ramanan, R.N.; Selvaraj, M.; Vijayaraghavan, R.; MacFarlane, D.R.; Ooi, C.W. Structural Stability of Insulin Aspart in Aqueous Cholinium Aminoate Ionic Liquids Based on Molecular Dynamics Simulation Studies. J. Mol. Liq. 2021, 322, 114501. [Google Scholar] [CrossRef]
- Banerjee, A.; Ibsen, K.; Iwao, Y.; Zakrewsky, M.; Mitragotri, S. Transdermal Protein Delivery Using Choline and Geranate (CAGE) Deep Eutectic Solvent. Adv. Healthc. Mater. 2017, 6, 1601411. [Google Scholar] [CrossRef] [Green Version]
- Jorge, L.R.; Harada, L.K.; Silva, E.C.; Campos, W.F.; Moreli, F.C.; Shimamoto, G.; Pereira, J.F.B.; Oliveira, J.M.; Tubino, M.; Vila, M.M.D.C.; et al. Non-Invasive Transdermal Delivery of Human Insulin Using Ionic Liquids: In Vitro Studies. Front. Pharmacol. 2020, 11, 243. [Google Scholar] [CrossRef] [Green Version]
- Hattori, T.; Tagawa, H.; Inai, M.; Kan, T.; Kimura, S.-I.; Itai, S.; Mitragotri, S.; Iwao, Y. Transdermal Delivery of Nobiletin Using Ionic Liquids. Sci. Rep. 2019, 9, 20191. [Google Scholar] [CrossRef]
- Banerjee, A.; Ibsen, K.; Brown, T.; Chen, R.; Agatemor, C.; Mitragotri, S. Ionic Liquids for Oral Insulin Delivery. Proc. Natl. Acad. Sci. USA 2018, 115, 7296–7301. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y.; Zhao, Z.; Gao, Y.; Pan, D.C.; Salinas, A.K.; Tanner, E.E.L.; Guo, J.; Mitragotri, S. Oral Delivery of Sorafenib through Spontaneous Formation of Ionic Liquid Nanocomplexes. J. Control. Release 2020, 322, 602–609. [Google Scholar] [CrossRef]
- Shi, Y.; Zhao, Z.; Peng, K.; Gao, Y.; Wu, D.; Kim, J.; Mitragotri, S. Enhancement of Anticancer Efficacy and Tumor Penetration of Sorafenib by Ionic Liquids. Adv. Healthc. Mater. 2021, 10, 2001455. [Google Scholar] [CrossRef]
- Jungwirth, P. Biological Water or Rather Water in Biology? J. Phys. Chem. Lett. 2015, 6, 2449–2451. [Google Scholar] [CrossRef]
- Bash, E. Water and Proteins: A Love-Hate Relationship Yaakov. Proc. Natl. Acad. Sci. USA 2015, 1, 3325–3326. [Google Scholar] [CrossRef]
- Tanner, E.E.L.; Piston, K.M.; Ma, H.; Ibsen, K.N.; Nangia, S.; Mitragotri, S. The Influence of Water on Choline-Based Ionic Liquids. ACS Biomater. Sci. Eng. 2019, 5, 3645–3653. [Google Scholar] [CrossRef]
- Palanisamy, K.; Prakash, M. The Molecular Mechanism behind the Stabilization of Insulin by Choline and Geranate (CAGE) Ionic Liquids-Computational Insights into Oral Insulin Drug Formulation. Phys. Chem. Chem. Phys. 2021, 23, 25298–25307. [Google Scholar] [CrossRef]
- Kao, W.H.L.; Puddey, I.B.; Boland, L.L.; Watson, R.L.; Brancati, F.L. Alcohol Consumption and the Risk of Type 2 Diabetes Mellitus: Atherosclerosis Risk in Communities Study. Am. J. Epidemiol. 2001, 154, 748–757. [Google Scholar] [CrossRef] [Green Version]
- Howard, A.A.; Arnsten, J.H.; Gourevitch, M.N. Effect of Alcohol Consumption on Diabetes Mellitus: A Systematic Review. Ann. Intern. Med. 2004, 140, 211–224. [Google Scholar] [CrossRef]
- Kim, S.J.; Kim, D.J. Alcoholism and Diabetes Mellitus. Diabetes Metab. J. 2012, 36, 108–115. [Google Scholar] [CrossRef]
- Ratha, B.N.; Kar, R.K.; Bednarikova, Z.; Gazova, Z.; Kotler, S.A.; Raha, S.; De, S.; Maiti, N.C.; Bhunia, A. Molecular Details of a Salt Bridge and Its Role in Insulin Fibrillation by NMR and Raman Spectroscopic Analysis. J. Phys. Chem. B 2020, 124, 1125–1136. [Google Scholar] [CrossRef]
- Banerjee, P.; Mondal, S.; Bagchi, B. Adverse Effect of Ethanol on Insulin Dimer Stability. arXiv 2018, arXiv:1809.00807. [Google Scholar]
- Mukherjee, S.; Deshmukh, A.A.; Mondal, S.; Gopal, B.; Bagchi, B. Destabilization of Insulin Hexamer in Water-Ethanol Binary Mixture. J. Phys. Chem. B 2019, 123, 10365–10375. [Google Scholar] [CrossRef]
- Banerjee, P.; Mondal, S.; Bagchi, B. Effect of Ethanol on Insulin Dimer Dissociation. J. Chem. Phys. 2019, 150, 084902. [Google Scholar] [CrossRef] [PubMed]
- Acharya, S.; Mondal, S.; Mukherjee, S.; Bagchi, B. Rate of Insulin Dimer Dissociation: Interplay between Memory Effects and Higher Dimensionality. J. Phys. Chem. B 2021, 125, 9678–9691. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Acharya, S.; Mondal, S.; Banerjee, P.; Bagchi, B. Structural Stability of Insulin Oligomers and Protein Association-Dissociation Processes: Free Energy Landscape and Universal Role of Water. J. Phys. Chem. B 2021, 125, 11793–11811. [Google Scholar] [CrossRef] [PubMed]
- Mondal, S.; Mukherjee, S.; Acharya, S.; Bagchi, B. Unfolding of Dynamical Events in the Early Stage of Insulin Dimer Dissociation. J. Phys. Chem. B 2021, 125, 7958–7966. [Google Scholar] [CrossRef] [PubMed]
- Zoete, V.; Meuwly, M.; Karplus, M. Study of the Insulin Dimerization: Binding Free Energy Calculations and per-Residue Free Energy Decomposition. Proteins: Struct. Funct. Genet. 2005, 61, 79–93. [Google Scholar] [CrossRef]
- Gong, Q.; Zhang, H.; Zhang, H.; Chen, C. Calculating the Absolute Binding Free Energy of the Insulin Dimer in an Explicit Solvent. RSC Adv. 2019, 10, 790–800. [Google Scholar] [CrossRef] [Green Version]
- Baker, E.N.; Blundell, T.L.; Cutfield, J.F.; Cutfield, S.M.; Dodson, E.J.; Dodson, G.G.; Hodgkin, D.M.; Hubbard, R.E.; Isaacs, N.W.; Reynolds, C.D. The Structure of 2Zn Pig Insulin Crystals at 1.5 A Resolution. Philos. Trans. R. Soc. Lond. B B Biol. Sci. 1988, 319, 369–456. [Google Scholar] [CrossRef]
- Mark, P.; Nilsson, L. Structure and Dynamics of the TIP3P, SPC, and SPC/E Water Models at 298 K. J. Phys. Chem. A 2001, 105, 9954–9960. [Google Scholar] [CrossRef]
- Lindorff-Larsen, K.; Piana, S.; Palmo, K.; Maragakis, P.; Klepeis, J.L.; Dror, R.O.; Shaw, D.E. Improved Side-Chain Torsion Potentials for the Amber Ff99SB Protein Force Field. Proteins Struct. Funct. Bioinform. 2010, 78, 1950–1958. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and Testing of a General Amber Force Field. J. Comput. Chem. 2004, 56531, 1157–1174. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle Mesh Ewald: An N·log(N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef] [Green Version]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A Smooth Particle Mesh Ewald Method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef] [Green Version]
- Van Gunsteren, W.F.; Berendsen, H.J.C. A Leap-Frog Algorithm for Stochastic Dynamics. Mol. Simul. 1988, 1, 173–185. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A Linear Constraint Solver for Molecular Simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- DeLano, W.L. Pymol: An Open-Source Molecular Graphics Tool. CCP4 Newsl. Protein Crystallogr. 2002, 40, 82–92. [Google Scholar]
- Ricker, E.W. VMD: Visual Molecular Dynamics. J. Fish. Res. Board Can. 1954, 11, 559–623. [Google Scholar] [CrossRef]
- Miller, B.R.; McGee, T.D.; Swails, J.M.; Homeyer, N.; Gohlke, H.; Roitberg, A.E. MMPBSA.Py: An Efficient Program for End-State Free Energy Calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef]
- Barducci, A.; Bussi, G.; Parrinello, M. Well-Tempered Metadynamics: A Smoothly Converging and Tunable Free-Energy Method. Phys. Rev. Lett. 2008, 100, 200–212. [Google Scholar] [CrossRef] [Green Version]
- Bussi, G.; Laio, A. Using Metadynamics to Explore Complex Free-Energy Landscapes. Nat. Rev. Phys. 2020, 2, 200–212. [Google Scholar] [CrossRef]
- Bonomi, M. The PLUMED consortium. Promoting transparency and reproducibility in enhanced molecular simulations. Nat. Methods 2019, 16, 670–673. [Google Scholar] [CrossRef]
- Takeda, J.; Iwao, Y.; Karashima, M.; Yamamoto, K.; Ikeda, Y. Structural Evaluation of the Choline and Geranic Acid/Water Complex by SAXS and NMR Analyses. ACS Biomater. Sci. Eng. 2021, 7, 595–604. [Google Scholar] [CrossRef] [PubMed]







| System | Mole Fraction of EtOH | EtOH | Choline | Geranate | Water |
|---|---|---|---|---|---|
| 1 | Ins-Water | 0 | 0 | 0 | 8229 |
| 2 | 0.10 | 30 | 270 | 270 | 3748 |
| 3 | 0.20 | 60 | 240 | 240 | 4111 |
| 4 | 0.40 | 120 | 180 | 180 | 4791 |
| 5 | 0.60 | 180 | 120 | 120 | 5595 |
| 6 | 0.80 | 240 | 60 | 60 | 6330 |
| 7 | 1.00 | 300 | 0 | 0 | 7183 |
| System | Mole Fraction of EtOH | EtOH | Choline | Geranate | Water | REtOH | RCholine | RGeranate |
|---|---|---|---|---|---|---|---|---|
| 1 | 0.10 | 2.79 | 27.60 | 36.27 | 256.15 | 1.35 | 1.49 | 1.96 |
| 2 | 0.20 | 6.93 | 25.09 | 31.56 | 279.39 | 1.71 | 1.54 | 1.93 |
| 3 | 0.40 | 16.83 | 21.96 | 33.34 | 274.84 | 2.44 | 2.13 | 3.23 |
| 4 | 0.60 | 20.20 | 11.87 | 19.64 | 218.12 | 2.88 | 2.54 | 4.20 |
| 5 | 0.80 | 28.13 | 6.86 | 13.84 | 237.49 | 3.12 | 3.06 | 6.19 |
| 6 | 1.00 | 38.66 | 00 | 00 | 297.66 | 3.11 | --- | --- |
| Mole Fractions of EtOH | Enthalpy (∆H) | Entropy (∆S) | Total Binding Free Energy (∆Gbind) |
|---|---|---|---|
| 0.10 | −49.69 | −36.94 | −12.75 |
| 0.20 | −50.98 | −34.88 | −16.1 |
| 0.40 | −49.32 | −41.08 | −8.24 |
| 0.60 | −47.62 | −33.88 | −13.74 |
| 0.80 | −50.22 | −29.72 | −20.5 |
| 1.00 | −50.51 | −38.50 | −12.01 |
| Ins-Water | −52.20 | −34.70 | −17.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Palanisamy, K.; Prakash, M. Counteractive Effects of Choline Geranate (CAGE) ILs and Ethanol on Insulin’s Stability—A Leap Forward towards Oral Insulin Formulation. Molecules 2022, 27, 5031. https://doi.org/10.3390/molecules27155031
Palanisamy K, Prakash M. Counteractive Effects of Choline Geranate (CAGE) ILs and Ethanol on Insulin’s Stability—A Leap Forward towards Oral Insulin Formulation. Molecules. 2022; 27(15):5031. https://doi.org/10.3390/molecules27155031
Chicago/Turabian StylePalanisamy, Kandhan, and Muthuramalingam Prakash. 2022. "Counteractive Effects of Choline Geranate (CAGE) ILs and Ethanol on Insulin’s Stability—A Leap Forward towards Oral Insulin Formulation" Molecules 27, no. 15: 5031. https://doi.org/10.3390/molecules27155031