Evaluation of the Antiviral Activity against Infectious Pancreatic Necrosis Virus (IPNV) of a Copper (I) Homoleptic Complex with a Coumarin as Ligand
Abstract
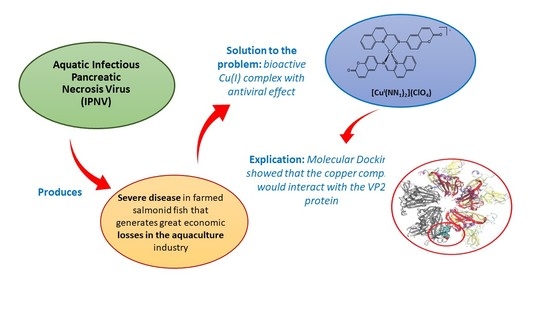
:1. Introduction
2. Results and Discussion
2.1. Chemical Compunds
2.2. Cytotoxicity Test
2.3. Antiviral Activity
2.4. Computational Studies
3. Materials and Methods
3.1. Cytotoxicity Test
3.2. Antiviral Activity Evaluation
3.2.1. IPNV Propagation
3.2.2. Antiviral Activity and Viral Load Assay
3.2.3. RNA Extraction and cDNA Synthesis
3.2.4. Real Time Quantitative PCR
3.3. Computational Studies
3.4. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Tapia, D.; Eissler, Y.; Reyes-Lopez, F.; Kuznar, J.; Yáñez, J. Infectious pancreatic necrosis virus in salmonids: Molecular epidemiology and host response to infection. Rev. Aquac. 2021, 1–19. [Google Scholar] [CrossRef]
- Mutoloki, S.; Jøssund, T.; Ritchie, G.; Munang’andu, H.; Evensen, O. Infectious Pancreatic Necrosis Virus Causing Clinical and Subclinical Infections in Atlantic Salmon Have Different Genetic Fingerprints. Front. Microbiol. 2016, 7, 1393–1402. [Google Scholar] [CrossRef] [Green Version]
- Tapia, D.; Barría, A.; Kuznar, J.; Yáñez, J.M. Comparison of mortality and viral load in rainbow trout (Oncorhynchus mykiss) infected with infectious pancreatic necrosis virus (IPNV) genogroups 1 and 5. J. Fish Dis. 2020, 43, 139–146. [Google Scholar] [CrossRef]
- Dopazo, C.P. The Infectious Pancreatic Necrosis Virus (IPNV) and its Virulence Determinants: What is Known and What Should be Known. Pathogens 2020, 9, 94. [Google Scholar] [CrossRef] [Green Version]
- Rivas-Aravena, A.; Martin, M.C.-S.; Galaz, J.; Imarai, M.; Miranda, D.; Spencer, E.; Sandino, A. Evaluation of the immune response against immature viral particles of infectious pan-creatic necrosis virus (IPNV): A new model to develop an attenuated vaccine. Vaccine 2012, 30, 5110–5117. [Google Scholar] [CrossRef]
- Dobos, P. Size and structure of the genome of infectious pancreatic necrosis virus. Nucleic Acids Res. 1976, 3, 1903–1924. [Google Scholar] [CrossRef] [Green Version]
- Duncan, R.; Nagy, E.; Krell, P.; Dobos, P. Synthesis of the infectious pancreatic necrosis vi-rus polyprotein, detection of a virus-encoded protease, and fine structure mapping of genome segment A coding regions. J. Virol. 1987, 61, 3655–3664. [Google Scholar] [CrossRef] [Green Version]
- Galloux, M.; Chevalier, C.; Henry, C.; Huet, J.-C.; Da Costa, B.; Delmas, B. Peptides resulting from the pVP2 C-terminal processing are present in infectious pancreatic necrosis virus particles. J. Gen. Virol. 2004, 85, 2231–2236. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, B.L. Use of monoclonal antibodies in identification and of fish viruses. Annu. Rev. Fish Dis. 1993, 3, 241–257. [Google Scholar] [CrossRef]
- Hjalmarsson, A.; Carlemalm, E.; Everitt, E. Infectious pancreatic necrosis virus: Identification of a VP3-containing ribonucleoprotein core structure and evidence for Olinked glycosylation of the capsid protein. J. Virol. 1999, 73, 3484–3490. [Google Scholar] [CrossRef] [Green Version]
- Ortega, C.; Enríquez, R. Factores asociados a la infección celular por el virus de la necrosis pancreática infecciosa (IPNV). Arch. Med. Veter. 2007, 39, 7–18. [Google Scholar] [CrossRef] [Green Version]
- Santi, N.; Song, H.; Vakharia, V.N.; Evensen, Ø. Infectious Pancreatic Necrosis Virus VP5 Is Dispensable for Virulence and Persistence Infectious Pancreatic Necrosis Virus VP5 Is Dispensable for Virulence and Persistence. J. Virol. 2005, 79, 9206–9216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dobos, P. The molecular biology of infectious pancreatic necrosis virus (IPNV). Annu. Rev. Fish Dis. 1995, 5, 25–54. [Google Scholar] [CrossRef]
- Duncan, R.; Mason, C.L.; Nagy, E.; Leong, J.; Dobos, P. Sequence analysis of infectious pancreatic necrosis virus genome segment B and its encoded VP1 protein: A putative RNA-dependent RNA polymerase lacking the Gly-Asp-Asp motif. Virology 1991, 181, 541–552. [Google Scholar] [CrossRef]
- Fridholm, H.; Eliasson, L.; Everitt, E. Immunogenicity properties of authentic and heterologously synthesized structural protein VP2 of infectious pancreatic necrosis virus. Viral Immunol. 2007, 20, 635–648. [Google Scholar] [CrossRef]
- Mileva, E. Infectious pancreatic necrosis of salmonid fish—Distribution and laboratory methods for diagnosis. Trakia J. Sci. 2019, 17, 401–412. [Google Scholar] [CrossRef]
- Julin, K.; Johansen, L.; Sommer, A.; Jørgensen, J. Persistent infections with infectious pancreatic necrosis virus (IPNV) of different virulence in Atlantic salmon, Salmo salar L. J. Fish Dis. 2014, 38, 1005–1019. [Google Scholar] [CrossRef]
- Nygaard, H.; Modahl, I.; Myrmel, M. Thermal Inactivation of Infectious Pancreatic Necrosis Virus in a Peptone-Salt Medium Mimicking the Water-Soluble Phase of Hydrolyzed Fish By-Products. Appl. Environ. Microbiol. 2012, 78, 2446–2448. [Google Scholar] [CrossRef]
- Iakovidis, I.; Delimaris, I.; Piperakis, S. Copper and Its Complexes in Medicine: A Biochemical Approach. Mol. Biol. Int. 2011, 2011, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Aldabaldetrecu, M.; Parra, M.; Soto, S.; Arce, P.; Tello, M.; Guerrero, J.; Modak, B. New Copper (I) Complex with a Coumarin as Ligand with Antibacterial Activity against Flavobacterium psychrophilum. Molecules 2020, 25, 3183. [Google Scholar] [CrossRef] [PubMed]
- Soto-Aguilera, S.; Modak, B.; Aldabaldetrecu, M.; Lozano, C.P.; Guerrero, J.; Lefimil, C.; Parra, M. In Vitro Effect of Copper (I) Complex [Cu(NN1)2](ClO4) on Vibrio harveyi BB170 Bio-film Formation. Microorganisms 2021, 9, 2273. [Google Scholar] [CrossRef]
- Singh, H.; Singh, J.V.; Bhagat, K.; Gulati, H.K.; Sanduja, M.; Kumar, N.; Kinarivala, N.; Sharma, S. Rational approaches, design strategies, structure activity relationship and mechanistic insights for therapeutic coumarin hybrids. Bioorg. Med. Chem. 2019, 27, 3477–3510. [Google Scholar] [CrossRef] [PubMed]
- Grosell, M. Homeostasis and Toxicology of Essential Metals. In Fish Physiology, 1st ed.; Chris, M., Wood, C., Farrell, A., Brauner, C., Eds.; Academic Press: Cambridge, MA, USA, 2012; Volume 31, Part A; pp. 53–133. [Google Scholar]
- Claudel, M.; Schwarte, J.; Fromm, K. New Antimicrobial Strategies Based on Metal Complexes. Chemistry 2020, 2, 849–899. [Google Scholar] [CrossRef]
- Kalinowska, M.; Piekut, J.; Bruss, A.; Follet, C.; Sienkiewicz-Gromiuk, J.; Świsłocka, R.; Rzączyńska, Z.; Lewandowski, W. Spectroscopic (FT-IR, FT-Raman, 1H, 13C NMR, UV/VIS), thermogravimetric and antimicrobial studies of Ca(II), Mn(II), Cu(II), Zn(II) and Cd(II) complexes of ferulic acid. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2014, 122, 631–638. [Google Scholar] [CrossRef]
- Lenard, J. Viral Membranes. Encyclopedia of Virology, 3rd ed.; Mahy, B.W.J., Van Regenmortel, M.H.V., Eds.; Academic Press: Cambridge, MA, USA, 2008; pp. 308–314. [Google Scholar] [CrossRef]
- Pereiro, P.; Figueras, A.; Novoa, B. Compilation of antiviral treatments and strategies to fight fish viruses. Rev. Aquac. 2021, 13, 1223–1254. [Google Scholar] [CrossRef]
- Song, H.; Santi, N.; Evensen, Ø.; Vakharia, V.N. Molecular determinants of Infectious Pancreatic Necrosis Virus virulence and cell culture adaptation. J. Virol. 2005, 79, 10289–10299. [Google Scholar] [CrossRef] [Green Version]
- Kuznar, J.; Soler, M.; Farias, G.; Espinoza, J.C. Attachment and entry of infectious pancreatic necrosis virus (IPNV) into CHSE-214 cells. Arch. Virol. 1995, 140, 1833–1840. [Google Scholar] [CrossRef]
- Modak, B.; Sandino, A.M.; Arata, L.; Cárdenas-Jirón, G.; Torres, R. Inhibitory effect of aromatic geranyl derivatives isolated from Heliotropium filifolium on infectious pancreatic necrosis virus replication. Veter. Microbiol. 2010, 141, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Dennington, R.; Keith, T.; May, J.M. GaussView; Version 5; Semichem Inc.: Shawnee, KS, USA, 2009. [Google Scholar]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 09; Gaussian, Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Coulibaly, F.; Chevalier, C.; Delmas, B.; Rey, F.A. Crystal Structure of an Aquabirnavirus Particle: Insights into Antigenic Diversity and Virulence Determinism. J. Virol. 2010, 84, 1792–1799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morris, G.M.; Goodsell, D.S.; Halliday, R.S.; Huey, R.; Hart, W.E.; Belew, R.K.; Olson, A.J. Automated Docking Using a Lamarckian Genetic Algorithm and an Empirical Binding Free Energy Function. J. Comput. Chem. 1998, 19, 1639–1662. [Google Scholar] [CrossRef] [Green Version]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kalé, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef] [Green Version]
- Berendsen, H.J.C.; Postma, J.P.M.; Van Gunsteren, W.F.; DiNola, A.; Haak, J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef] [Green Version]




| Target | Primer Sequence (5′-3′) | Accession Number |
|---|---|---|
| VP2 | ACCAAGTTCGACTTCCAGC ATCGGCTTGGTGATGTTCTC | GenBank: FN257531.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gutiérrez, D.; Benavides, A.; Valenzuela, B.; Mascayano, C.; Aldabaldetrecu, M.; Olguín, A.; Guerrero, J.; Modak, B. Evaluation of the Antiviral Activity against Infectious Pancreatic Necrosis Virus (IPNV) of a Copper (I) Homoleptic Complex with a Coumarin as Ligand. Molecules 2022, 27, 32. https://doi.org/10.3390/molecules27010032
Gutiérrez D, Benavides A, Valenzuela B, Mascayano C, Aldabaldetrecu M, Olguín A, Guerrero J, Modak B. Evaluation of the Antiviral Activity against Infectious Pancreatic Necrosis Virus (IPNV) of a Copper (I) Homoleptic Complex with a Coumarin as Ligand. Molecules. 2022; 27(1):32. https://doi.org/10.3390/molecules27010032
Chicago/Turabian StyleGutiérrez, Daniela, Almendra Benavides, Beatriz Valenzuela, Carolina Mascayano, Maialen Aldabaldetrecu, Angel Olguín, Juan Guerrero, and Brenda Modak. 2022. "Evaluation of the Antiviral Activity against Infectious Pancreatic Necrosis Virus (IPNV) of a Copper (I) Homoleptic Complex with a Coumarin as Ligand" Molecules 27, no. 1: 32. https://doi.org/10.3390/molecules27010032