Membrane proteins are important drug targets, which make screening of chemical libraries and studies of drug binding to these targets crucial for drug discovery and structure-based drug design approaches. Since solution NMR is not accessible to protein-membrane complexes due to the large size (>MDa) one can apply solid-state NMR. However, the resolution and sensitivity is quite limited compared to the solution NMR. Therefore, we use here intact Bcl-2 and variants retaining specific functional subunits in membrane-mimicking micellar environments which are accessible to solution NMR to obtain high-quality NMR spectra of Bcl-2 while retaining its function. Large membrane proteins with multiple domains and large loop regions also suffer from poor resolution due to peak overlap. Therefore, optimization of detergents, temperature, and protein sequence length are required for solution membrane protein NMR drug discovery approaches.
2.1. NMR Spectra of Bcl-2 Protein: Choice of Micellar System and Temperature
The spectral quality of NMR is critically dependent on the size and the tumbling rate of the studied system. Therefore, it is important to find a combination of membrane proteins and detergent which produce good sensitivity and spectral resolution while keeping the main functional feature of the protein intact. We tested a broad range of detergents as solubilization agents for the full length and truncated variants of the Bcl-2 protein, based on our previous work with cell-free expression systems. Out of this detergent screens which started with the detergent systems Brij 35 (starting point) the best outcome was DPC (Dodecylphosphocholine) with the result seen in
Figure 1. Brij 35 itself is a non-ionic detergent with MW 1200, aggregation number(n) 40, CMC and micellar-size around < 0.1 mM and 48 kDa respectively. While DPC is a zwitterionic headgroup detergent with MW 351, aggregation number(n) 54, CMC and micellar-size around 1.5 mM and 19 kDa respectively [
20,
21]. As the starting point we used Brij-35 detergent, which turned out to be ideal for purification of intact human Bcl-2 protein by cell-free and standard E. coli overexpression systems [
17,
22] prior any further detergent exchange e.g., in DPC micelles. In those micellar environments, the Bcl-2 protein was fully functional as shown by a very high affinity constant (K
D around 35.8 nM) upon titration of its natural inhibiting partner, the soluble apoptotic Bax-protein [
16]. This detergent system also enables the proper reconstitution of intact Bcl-2 protein back into its native lipid membrane environment.
However, as clearly seen here in
Figure 1A, the corresponding
1H-
15N-TROSY NMR spectrum of Bcl-2 in Brij-35 micelles at 298 K, reveals only a fraction of peaks well resolved (ca. 52 peaks) while the remaining ones are broadened or undetectable due to the slow tumbling rate of quite large Brij-35 micelles. Therefore, the spectral quality is not sufficient to identify most resonances of the Bcl-2 protein, which also makes it insufficient for any NMR-based structural work. Only the sharp resonances which belong to the extended (32–90) intrinsically disordered loop region FLD (as shown further down) can be clearly identified in the spectrum as separate peaks. By changing the detergent system to DPC which forms smaller micelles with faster tumbling rates a different picture is emerging. As seen in
Figure 1A, the NMR spectra of the Bcl-2/DPC system reveal many more identifiable peaks (ca. 165 peaks) compared to Brij-35 under similar conditions. Clearly, the smaller size of the micellar systems with its increased tumbling rate provides significant advantages to investigate larger membrane proteins in a membrane-mimicking micellar environment by solution NMR methods. In the corresponding
1H-
15N-TROSY NMR spectrum (
Figure 1A, red) not only the peaks of the FLD domain are visible but also most resonances of the main folded domains composed of the N-terminal head (BH4 + BH3-1 domain). Deuteration of the protein did increase this number slightly compared to the non-deuterated Bcl-2 protein at 298 K, but not at higher temperatures, which improved both sensitivity (due to fast tumbling) and peak resolution of spectra for non-deuterated Bcl-2 with an optimum at 328 K. For non-deuterated Bcl-2 at the physiological temperature (310 K) and above at 328 K around 170 and 190 backbone signals were observed in the
1H-
15N-TROSY spectrum respectively. Since full-length Bcl-2 protein consists of 25% of residues belonging to it intrinsically disordered FLD sequence, signals from this FLD region overlap with signals from the remaining protein causing severe peak overlapping in the 2D
1H-
15N TROSY spectra. Combined with experiments at physiological temperatures it creates a bottleneck for structure-characterization due to the less sensitive
13C Cβ/Cα signal-intensities (for membrane-embedded BH1-3 domains) in 3D experiments. Therefore, increasing the temperature to 328 K (which increases tumbling rates for the temperature-stable Bcl-2 micelle system), proved advantageous since it provides a good signal to noise ratio of
13C signals from the side-chains of amino acids in the triple-resonance experiments.
Another important parameter to improve the spectral quality of Bcl-2 in DPC micelles is temperature. By increasing the temperature the overall tumbling rate becomes faster, providing better resolved NMR spectra. In addition, the increase of temperature also has a positive effect on the molecular dynamics of chemical moieties which reside on an intermediate NMR time scale at lower temperature, often connected with broadened or even non-detectable resonances. Moving those motional processes (e.g., local exchange or wobbling of molecular segments) into the fast region of the NMR time scale produces well resolved NMR spectra. As seen in
Figure 1B, by increasing in temperature, the number of NMR peaks in the
1H-
15N-TROSY spectrum also increases, with each peak representing one of the maximum 239 aa in the human Bcl-2 construct. Since rising temperature causes a faster tumbling rate of protein: micellar complexes, a comparison is shown in
Figure 1B for temperatures 298 K, 310 K, and 328 K. Bcl-2 protein in 5 mM DPC buffer was stable at 328 K for nearly two weeks and upon cooling to 310 K or 298 K similar spectra were produced indicating that Bcl-2 protein in DPC micelles can maintain its stability above the physiological temperatures required for structure characterization with solution state NMR.
At higher or above physiological temperatures (>310 K), Bcl-2 or its truncated variants retain the folded structures in DPC micelles observed by the temperature dependent Far-UV Circular Dichroism (CD) measurements (
Supplementary Figure S1A–E). Also, comparing the
1H-
15N-TROSY spectra of Bcl-2 at 5 mM DPC (above CMC) conditions with the ones with below CMC at 0.25 mM DPC (see
Supplementary Figure S2), indicate that the protein is still folded below the CMC of the detergent, thus generating very similar spectra. Refolded Bcl-2 or the truncated variants hold large amounts of DPC molecules.
31P NMR spectra of refolded Bcl-2 or the truncated variants in 5 mM DPC micelles in NMR buffer supports this observation when comparing spectra with and without DPC present (
Supplementary Figure S3A,B). This strong lipid affinity toward the Bcl-2 structure is very important in the process of the MOM insertion. The embedded Bcl-2 state can be observed with NMR starting with the soluble fraction of Bcl-2 ∆TM. With
1H-
15N-TROSY-HSQC spectra we observe the structural transition in Bcl-2 from a soluble to membrane-mimic embedded state, when the soluble fraction of Bcl-2 ΔTM was treated with different concentrations of DPC to mimic the membrane environments. At 0.5 mM DPC there were only specific local changes in the protein backbone and tryptophan side-chain
15Nε-
1Hε cross-peaks (
Supplementary Figure S4B). At 10 mM DPC, an increase in the concentration from below CMC to above CMC, significant changes were observed globally in the protein, and the tryptophan side-chain
15Nε-
1Hε cross-peaks show a pattern similar to the refolded Bcl-2 ∆TM in 5 mM DPC buffer (
Supplementary Figure S4A,B), indicating a structural-transition from a soluble to membrane-embedded state. We performed the functional assay of Bcl-2 and Bcl-2 truncated variants (Bcl-2 ∆TM, Bcl-2 ∆N(1–82), and Bcl-2 ∆N(1–82) ∆TM(208–239)) with conserved BH3-1 domain in membrane-mimicking environments using the peptide from the BH3 domain of Bax. Binding titrations were performed under similar buffer and experiment conditions to investigate the chemical shift perturbations (CSP’s) due to peptide binding. Binding titration results show that the BH3-1 domain is important for BH3 peptide binding. In addition a comparison between the binding of Bcl-2 ∆N(1–82) and Bcl-2 ∆N(1–82) ∆TM(208–239) shows that the TM domain also contributes to the binding affinity. (
Supplementary Figure S5A–D). In
Supplementary Figure S5E, CSP of backbone amide (
1H,
15N and weighted
1H-
15N) along the Bax peptide molar ratio is plotted for three peaks 1-3 (shown as boxes in
Supplement Figure S5A,C) to highlight the differences upon binding titrations of full-length Bcl-2 and Bcl-2 ∆N(1–82). We observe similar patterns for the Bax peptide bindings in full-length Bcl-2 and Bcl-2 ∆N(1–82); however due to severe peak-overlaps in full-length Bcl-2 analysis becomes more difficult, while titrating the Bcl-2 ∆N(1–82) construct titration yields more information as there are no severe peak-overlaps. Plots of CSP along the Bax peptide molar ratio for the few backbone residues (peaks 1–3) show similar binding patterns with small differences in the affinities. However, the dynamic interplay or coordination between BH4 and BH3-1 TM domains via the FLD which has important significance for binding and inhibiting apoptotic full-length Bax can only be observed in full length Bcl-2.
2.2. The Effect of Bcl-2 Truncation on NMR Spectra
Since the full length 26kDa Bcl-2 protein is insoluble, a complete peak identification based solely on solution NMR spectra upon solubilization into micelles is still not straightforward. This is partially due to the micellar size but also that the Bcl-2 protein is composed of functional domains that generate partially overlapping NMR signals. Therefore, we have developed a “divide-and-conquer” strategy, where we produced constructs of Bcl-2 where specific domains and their functions were removed while the main protein core—its BH3-BH1 groove responsible for the sequestering of apoptotic Bcl-2 proteins—remained intact.
This strategy is based on our concept of Bcl-2 functioning based on the previous findings [
23,
24,
25] as outlined below and schematically visualized in
Figure 2.
Truncated Bcl-2 constructs with its extended FLD domain and hydrophobic TM-helix removed were shown to be soluble [
14]. The Bcl-2 ∆TM construct with its hydrophobic TM-helix removed is partially soluble. This soluble escape (partial solubility) of Bcl-2 ∆TM can be explained based on the crystal structures of truncated soluble Bcl-2 variants where BH4 and BH3-1 domains are non-covalently packed while the FLD or truncated loop is extended to the outside of Bcl-2, and is also seen in other systems such as Bcl-xL [
13]. Upon membrane insertion, the Bcl-2 domain organization based on the functions are shown as N-terminal BH4, FLD, and C-terminal BH3-1 and TM domains. N-terminal domains are known to interact with IP
3R and Raf-1 proteins [
23,
24,
25], while C-terminal BH3-1 domains are known to interact with BH3-only proteins, which are linked to apoptosis.
Here we have now dissected Bcl-2 into constructs with either the TM domain missing (Bcl-2 ∆TM) or the BH4 + FLD region (Bcl-2 ∆N(1–82)). As seen in
Figure 3A, Bcl-2 contains a long regulative flexible loop domain (FLD; aa 32–90) which shows overlapping peaks in the
1H-
15N-TROSY spectrum with resonances from the protein’s main core body (BH3-1 and TM). By comparing the spectrum with the one obtained for Bcl-2 (
Figure 1A, red) with the Bcl-2 ∆N(1–82) version, one can clearly identify that these flexible N-terminal residues 1–90 comprise the most and intense signals in the spectrum of the full length Bcl-2 variant (grey). Most of those resonances originate from the loop region and the others from the N-terminal BH4 region attached to it. Most other NMR signals belong to the main protein body (93–239 aa) and are weaker and often broadened due to large molecular weight of the Bcl-2/DPC micellar complex. Clearly, the truncation of N-terminal loop residues (1–82) or ∆N(1–82) results in less overlapping NMR spectra with similar peak patterns as the full Bcl-2 at 328 K. Therefore, truncation of the N-terminal loop residues (1–82) improves the spectral quality required for structure characterization of full Bcl-2 and probes structural changes upon interaction with other partners, ranging from BH3-domains of apoptotic proteins to promising drug targets. Since the TM domain remains, the functionality of the main protein fold is not compromised since this domain still contributes to the high affinity of the hydrophobic groove region [
28].
Nevertheless also Bcl-2 without its TM domain is still functional as shown previously [
28], albeit with less affinity toward BH3 domains compared to full-length Bcl-2 (our observations). As seen in
Figure 3B (grey) the NMR spectrum of Bcl-2 ∆TM still shows an impact on the globular fold of the folded protein like the full-length one. Adding its spectrum together with the one obtained for Bcl-2 ∆N(1–82) (
Figure 3B (red)) provides in summary an overlaid spectrum which is identical to the one obtained from the entire Bcl-2 protein itself (
Figure 3A (grey). This clearly supports our “divide and conquer” strategy that dissecting Bcl-2 retains the structural features while keeping the specific functions intact. This is ideally not only for structural work of Bcl-2 in its membrane-mimicking micellar environment but also in the hunt for drug candidates specifically targeting one of those functional domains.
2.3. Domain Organization of Full-Length Bcl-2 under Membrane-Mimicking Conditions
Based on the functions of Bcl-2 in the membrane the Bcl-2 sequence is divided into an N-terminal BH4, FLD (aa 32–90) domain, and a C-terminal BH3-1 TM domain as shown in
Figure 4 (bottom) and
Supplementary Figure S6. N-terminal domains are known to interact with IP
3R, Raf-1 [
23,
25] while C-terminal BH3-1 domains are known to interact with BH3-only proteins. Using Bcl-2 truncated variants together with NMR we concluded that N-terminal BH4, FLD, and the C-terminal BH3-1 TM domains are independent, since Bcl-2 ∆N(1–82) and Bcl-2 ∆C (93–239) together produce the full-length Bcl-2
1H-
15N-TROSY spectrum, shown in
Figure 4 (top).
Since the full length Bcl-2 is insoluble while Bcl-2 ∆TM with the truncated transmembrane-helix is partially soluble, there is a soluble escape of Bcl-2 ∆TM where the truncated variant with its BH4 and BH3-1 domains are non-covalently fused with an extended flexible FLD or truncated loop. Further truncation of the N-terminal BH4 domain in Bcl-2 ∆TM also generated an insoluble Bcl-2 ∆N(1–82) ∆TM(208–239) construct (
Supplementary Figure S6A,B). Therefore, using NMR we can target both functional sites of Bcl-2 for anti-cancer drug design, mimicking the membrane environments which is not possible with truncated soluble structures of Bcl-2 where the N-terminal BH4 helix is fused in the globular fold with the TM domain being absent. Those versions cannot probe independent functions of N-terminal BH4 and flexible loop domains (
Supplementary Figure S6A).
This might be the solubility mechanism in cytosolic Bcl-2 homologues. When we further truncate the N-terminal BH4 domain in Bcl-2 ∆TM, the Bcl-2 ∆N(1–82) ∆TM construct was also insoluble (
Supplementary Figure S6A,B), which shows that packing of the BH4 domain to the BH3-1 domain assembly is important for the proper formation of the globular fold of Bcl-2. Therefore, using intact Bcl-2 in membrane-like micellar environments is an ideal high-value cancer target where both the N- and C-terminal binding sites can be screened for promising drug candidates via high throughput screening methods based on solution NMR, where suitable substance libraries with the main principles are outlined under paragraph 2.4.
2.4. Drug Discovery: NMR Screening of Intact Bcl-2 Protein against Compound Libraries
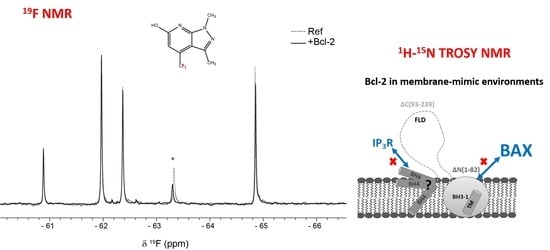
To provide new input into cancer therapy with Bcl-2 as the primary therapeutic target, fragment-based screening (FBS) using
19F NMR spectroscopy was performed.
19F NMR has several attractive properties compared to the more established
1H NMR used in FBS. Since the fluorine nucleus has a wide range of chemical shift values depending on its close molecular environment, it is normally not a problem to use mixtures of 10 or more fragments. Since those fragments are relatively simple and contain normally only one
19F nucleus, the individual fragments are easily identifiable in the corresponding spectra. Therefore, binding of a fragment to its target protein can simply be monitored as seen in
Figure 5. Another advantage is that the NMR experiments can be performed in non-deuterated organic buffers. Moreover, binding response in T
2-relaxation edited spectra are usually larger for
19F NMR because of the large chemical shift anisotropy of the
19F nucleus and a large line-width contribution from chemical exchange [
29].
Binding to Bcl-2 was investigated using T2-relaxation edited experiments of fragment mixtures in absence and presence of Bcl-2 where a reduction in peak intensity was observed for fragments binding to the protein as a result of fast exchange between free and bound form. The samples recorded in absence of Bcl-2 still contained DPC micelles to detect fragments binding directly to the DPC micelle surface; fragments that otherwise would be detected as false positives.
The commercially available Bionet fragment library, consisting of 428 fluorinated compounds was used in this study and screened in mixtures of 9–11 fragments per sample. Eight binding fragments were found, corresponding to a hit rate of approximately 2% (
Supplementary Table S1). In addition, 11 fragments bound to the micelle surface as evident by no or very small
19F signal intensity in the reference samples. A common motif observed in 3 of the 8 binding fragments consists of a disubstituted pyridine ring with a CF
3-group in the para position, see
Figure 6. This scaffold could potentially be an interesting starting point for structure—activity relationship studies.
Hits will be further assessed for their potential by validating their binding and determining the affinities using biophysical methods (ITC, TSA) and by NMR on
15N labelled protein. As shown in
Figure 3 and
Figure 4, 2D
1H-
15N NMR spectra can monitor binding of the molecule, its binding kinetics, and provide apparent binding affinities. Since each peak reflects a specific residue and its local environment we can assess if a binding event affects locally only the binding site (s. e.g.,
Figure 4) or if the entire protein undergoes structural rearrangements upon binding as seen in
Figure 3. Here, the BH3 peptide has most effect on the residues directly involved in its binding (s. also inserts in
Figure 3) but also the entire protein responds structurally (as seen by shifted resonances) to this binding event. The most affected residues presumably (ongoing assignment to confirm) belong to the Bcl-2′s binding groove (hotspot) and can be used to provide apparent affinities for a ligand upon titration; a strategy we will also exploit for new drug compounds to see if they interact to this key binding site of Bcl-2.