Transamination-Like Reaction Catalyzed by Leucine Dehydrogenase for Efficient Co-Synthesis of α-Amino Acids and α-Keto Acids
Abstract
:1. Introduction
2. Results and Discussion
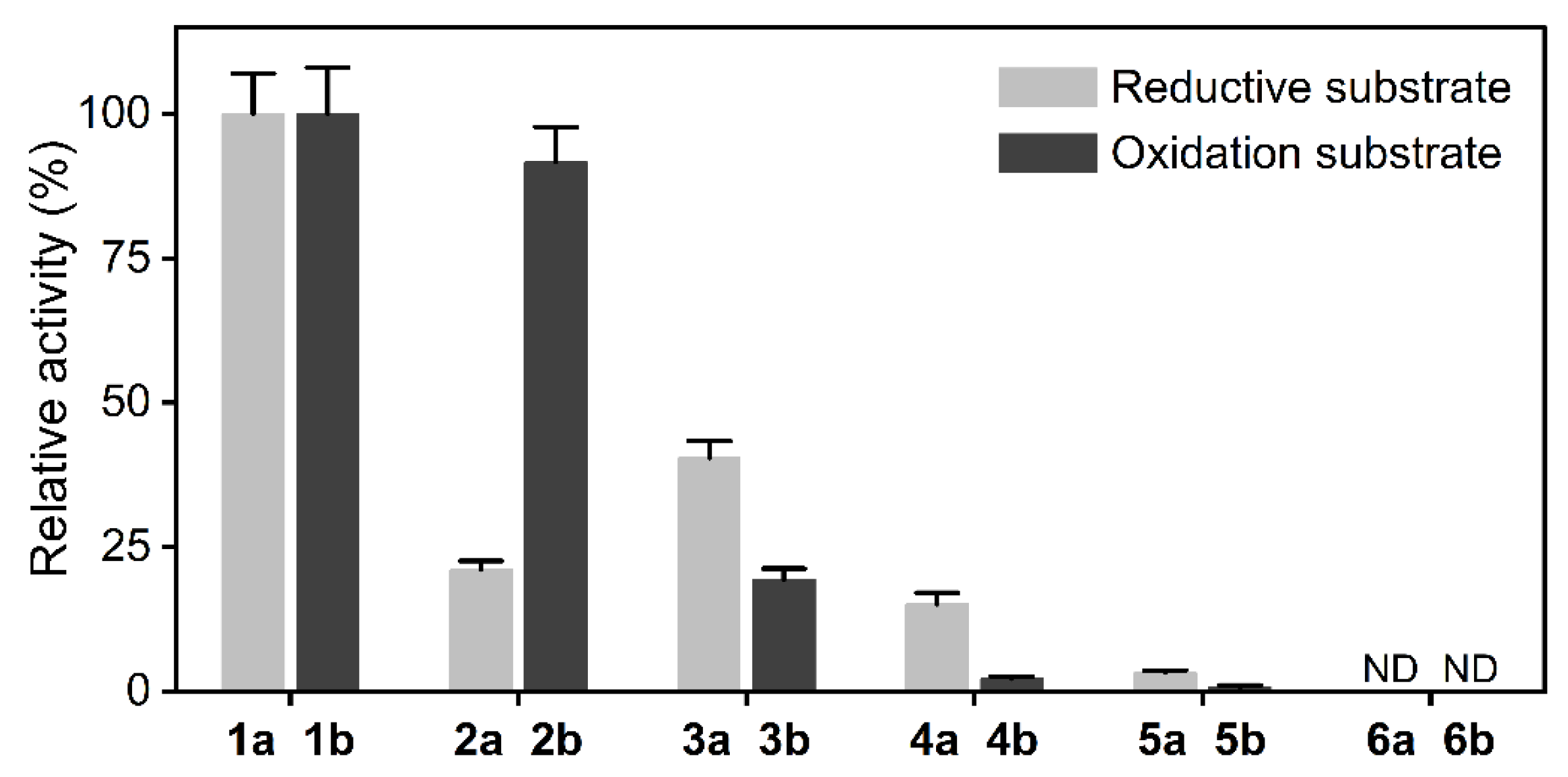
2.1. Substrate Specificity of LDH toward α-Amino Acids and α-Keto Acids
2.2. Conversion of Single-Step Oxidation and Reduction Reactions Catalyzed by LDH
2.3. Effect of Different Combinations of Amino Group Donor and Acceptor on Conversion Rate
2.4. Kinetic Parameters of LDH on Different Substrate
2.5. Optimization of Conditions for the Transamination-Like Reaction
2.6. Shifting Reaction Equilibrium by Adjusting the Substrate Ratio
3. Materials and Methods
3.1. Strains, Plasmids, and Chemicals
3.2. Protein Expression and Purification
3.3. Enzyme Activity Assay
3.4. Kinetic Parameters Analysis
3.5. Reaction Conditions Optimization
3.6. Reaction Progress Monitoring
3.7. HPLC Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Najera, C.; Sansano, J.M. Catalytic asymmetric synthesis of alpha-amino acids. Chem. Rev. 2007, 107, 4584–4671. [Google Scholar] [CrossRef]
- Basak, S.; Nader, S.; Mansy, S.S. Protometabolic reduction of NAD(+) with alpha-keto acids. JACS Au. 2021, 1, 371–374. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Xu, X.; Gao, C.; Zhang, Y.; Wu, J.; Liu, J.; Chen, X.; Luo, Q.; Liu, L. Open gate of corynebacterium glutamicum threonine deaminase for efficient synthesis of bulky alpha-keto acids. ACS Catal. 2020, 10, 9994–10004. [Google Scholar] [CrossRef]
- Galkin, A.; Kulakova, L.; Yoshimura, T.; Soda, K.; Esaki, N. Synthesis of optically active amino acids from α-keto acids with Escherichia coli cell expressing heterologous genes. Appl. Environ. Microbiol. 1997, 63, 4651–4656. [Google Scholar] [CrossRef] [Green Version]
- Penteado, F.; Lopes, E.F.; Alves, D.; Perin, G.; Jacob, R.G.; Lenardao, E.J. Alpha-keto acids: Acylating agents in organic synthesis. Chem. Rev. 2019, 119, 7113–7278. [Google Scholar] [CrossRef] [PubMed]
- Chun, S.W.; Narayan, A.R.H. Biocatalytic, stereoselective deuteration of alpha-amino acids and methyl esters. ACS Catal. 2020, 10, 7413–7418. [Google Scholar] [CrossRef] [PubMed]
- Moriwaki, H.; Resch, D.; Li, H.; Ojima, I.; Takeda, R.; Luis Acena, J.; Soloshonok, V. Inexpensive chemical method for preparation of enantiomerically pure phenylalanine. Amin. Acids. 2014, 46, 945–952. [Google Scholar] [CrossRef]
- Ogo, S.; Uehara, K.; Abura, T.; Fukuzumi, S. pH-dependent chemoselective synthesis of alpha-amino acids. Reductive amination of alpha-keto acids with ammonia catalyzed by acid-stable iridium hydride complexes in water. J. Am. Chem. Soc. 2004, 126, 3020–3021. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.Y.; Ji, Y.M.; Yu, Y.K.; Yu, J.Y.; Lee, Y.; Leet, S.J. Synthesis of alpha-ketobutyrolactones and gamma-hydroxy-alpha-keto acids. Bull. Korean Chem Soc. 2003, 24, 1819–1826. [Google Scholar]
- Xie, Y.; Lou, R.; Zhi, L.; Mi, A.; Jiang, Y. DPAMPP in catalytic asymmetric reactions: Enantioselective synthesis of L-homophenylalanine. Tetrahedron Asymmetry 2000, 11, 1487–1494. [Google Scholar] [CrossRef]
- Xue, Y.-P.; Cao, C.-H.; Zheng, Y.-G. Enzymatic asymmetric synthesis of chiral amino acids. Chem. Soc. Rev. 2018, 47, 1516–1561. [Google Scholar] [CrossRef]
- Simon, R.C.; Mutti, F.G.; Kroutil, W. Biocatalytic synthesis of enantiopure building blocks for pharmaceutical. Drug Discov. Today. 2013, 10, e37–e44. [Google Scholar] [CrossRef] [PubMed]
- Turner, N.J.; O’Reilly, E. Biocatalytic retrosynthesis. Nat. Chem. Biol. 2013, 9, 285–288. [Google Scholar] [CrossRef] [PubMed]
- Bornscheuer, U.T.; Huisman, G.W.; Kazlauskas, R.J.; Lutz, S.; Moore, J.C.; Robins, K. Engineering the third wave of biocatalysis. Nature 2012, 485, 185–194. [Google Scholar] [CrossRef]
- Bornscheuer, U.T. Biocatalysis: Successfully crossing boundaries. Angew. Chem. Int. Ed. 2016, 55, 4372–4373. [Google Scholar] [CrossRef] [Green Version]
- Yi, D.; Bayer, T.; Badenhorst, C.P.S.; Wu, S.; Doerr, M.; Höhne, M.; Bornscheuer, U.T. Recent trends in biocatalysis. Chem. Soc. Rev. 2021, 50, 8003–8049. [Google Scholar] [CrossRef]
- Sheldon, R.A.; Woodley, J.M. Role of biocatalysis in sustainable chemistry. Chem. Rev. 2018, 118, 801–838. [Google Scholar] [CrossRef] [PubMed]
- Savile, C.K.; Janey, J.M.; Mundorff, E.C.; Moore, J.C.; Tarn, S.; Jarvis, W.R.; Colbeck, J.C.; Krebber, A.; Fleitz, F.J.; Brands, J. Biocatalytic asymmetric synthesis of chiral amines from ketones applied to sitagliptin manufacture. Science 2010, 329, 305–309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slabu, I.; Galman, J.L.; Lloyd, R.C.; Turner, N.J. Discovery, engineering, and synthetic application of transaminase biocatalysts. ACS Catal. 2017, 7, 8263–8284. [Google Scholar] [CrossRef]
- Truppo, M.D.; Rozzell, J.D.; Moore, J.C.; Turner, N.J. Rapid screening and scale-up of transaminase catalysed reactions. Org. Biomol. Chem. 2009, 7, 395–398. [Google Scholar] [CrossRef]
- Truppo, M.D.; Strotman, H.; Hughes, G. Development of an immobilized transaminase capable of operating in organic solvent. ChemCatChem 2012, 4, 1071–1074. [Google Scholar] [CrossRef]
- Guo, F.; Berglund, P. Transaminase biocatalysis: Optimization and application. Green Chem. 2017, 19, 333–360. [Google Scholar] [CrossRef] [Green Version]
- Koike, K.; Hakamada, Y.; Yoshimatsu, T.; Kobayashi, T.; Ito, S. NADP-specific glutamate dehydrogenase from Alkaliphilic Bacillus sp. KSM-635: Purification and enzymatic properties. Biosci. Biotechnol. Biochem. 1996, 60, 1764–1767. [Google Scholar] [CrossRef] [Green Version]
- Li, H.M.; Zhu, D.M.; Hyatt, B.A.; Malik, F.M.; Biehl, E.R.; Ling, H. Cloning, protein sequence clarification, and substrates specificity of a leucine dehydrogenase from Bacillus sphaericus ATCC4525. Appl. Biochem. Biotechnol. 2009, 158, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Asano, Y.; Yamada, A.; Kato, Y.; Yamaguchi, K.; Hibino, Y.; Hirai, K.; Kondo, K. Enantioselective synthesis of (S)-amino acids by phenylalanine dehydrogenase from Bacillus sphaericus: Use of aatural and recombinant enzymes. J. Org. Chem. 1990, 55, 5567–5571. [Google Scholar] [CrossRef]
- Ouyang, S.; Li, X.; Sun, X.; Ouyang, J.; Yong, Q. A thermostable leucine dehydrogenase from Bacillus coagulans NL01: Expression, purification and characterization. Process. Biochem. 2020, 90, 89–96. [Google Scholar] [CrossRef]
- Yousefi, F.; Ataei, F.; Arab, S.S.; Hosseinkhani, S. Increase of Bacillus badius phenylalanine dehydrogenase specificity towards phenylalanine substrate by site-directed mutagenesis. Arch. Biochem Biophys. 2017, 635, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.-M.; Cheng, F.; Fu, F.-T.; Hu, H.-F.; Zheng, Y.-G. Semi-rational engineering of leucine dehydrogenase for L-2-aminobutyric acid production. Appl. Biochem. Biotechnol. 2017, 182, 898–909. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.P.; Wang, Y.L.; Chen, J.J.; Xu, M.J.; Yang, T.W.; Zheng, J.X.; Zhang, X.; Rao, Z.M. Rational engineering of Bacillus cereus leucine dehydrogenase towards α-keto Acid reduction for improving unnatural amino acid production. Biotechnol. J. 2019, 14, 1800253. [Google Scholar] [CrossRef]
- Zhou, F.; Mu, X.; Nie, Y.; Xu, Y. Enhanced catalytic efficiency and coenzyme affinity of leucine dehydrogenase by comprehensive screening strategy for L-tert-leucine synthesis. Appl. Microbiol. Biotechnol. 2021, 105, 3625–3634. [Google Scholar] [CrossRef]
- Luo, W.; Zhu, J.; Zhao, Y.; Zhang, H.; Yang, X.; Liu, Y.; Rao, Z.; Yu, X. Cloning and expression of a novel leucine dehydrogenase: Characterization and L-tert-leucine production. Front. Bioeng. Biotechnol. 2020, 8, 186. [Google Scholar] [CrossRef]
- Yin, X.J.; Liu, Y.Y.; Meng, L.J.; Zhou, H.S.; Wu, J.P.; Yang, L.R. Rational molecular engineering of glutamate dehydrogenases for enhancing asymmetric reductive amination of bulky α-keto acids. Adv. Synth. Catal. 2019, 361, 803–812. [Google Scholar] [CrossRef]
- Wu, T.; Mu, X.; Xue, Y.; Xu, Y.; Nie, Y. Structure-guided steric hindrance engineering of Bacillus badius phenylalanine dehydrogenase for efficient L-homophenylalanine synthesis. Biotechnol. Biofuels. 2021, 14, 207. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhu, R.; Zhou, J.; Yang, T.; Zhang, X.; Xu, M.; Rao, Z. Efficient single whole-cell biotransformation for L-2-aminobutyric acid production through engineering of leucine dehydrogenase combined with expression regulation. Bioresour Technol. 2021, 326, 124665. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.-Z.; Chang, D.-L.; Zhang, J. Discovery and application of new bacterial strains for asymmetric synthesis of L-tert-butyl leucine in high enantioselectivity. Appl. Biochem. Biotechnol. 2011, 164, 376–385. [Google Scholar] [CrossRef]
- Krishnamurthy, T. Chirality in microcystins. J. Am. Soc. Mass Spectrom. 1994, 5, 724–730. [Google Scholar] [CrossRef] [Green Version]
- B’Hymer, C.; Montes-Bayon, M.; Caruso, J.A. Marfey’s reagent: Past, present, and future uses of 1-fluoro-2,4-dinitrophenyl-5-L-alanine amide. J. Sep. Sci. 2003, 26, 7–19. [Google Scholar] [CrossRef]






| Substrate | Km (mM) | kcat (s−1) | kcat/Km (mM·s−1) |
|---|---|---|---|
| 1a | 1.16 ± 0.37 | 167.25 ± 10.23 | 143.69 ± 6.36 |
| 2a | 1.41 ± 0.19 | 73.20 ± 4.17 | 51.92 ± 1.57 |
| 3a | 2.38 ± 0.54 | 62.29 ± 4.32 | 26.14 ± 0.58 |
| 4a | 9.15 ± 1.23 | 16.79 ± 2.56 | 1.84 ± 0.32 |
| 1b | 0.86 ± 0.21 | 0.25 ± 0.03 | 0.29 ± 0.03 |
| 2b | 2.31 ± 0.11 | 0.17 ± 0.01 | 0.07 ± 0.001 |
| 3b | 30.04 ± 1.39 | 0.16 ± 0.06 | 0.005 ± 0.001 |
| 4b | 16.26 ± 0.86 | 0.05 ± 0.01 | 0.003 ± 0.002 |
| NAD+ | 0.25 ± 0.04 | 1.07 ± 0.249 | 2.08 ± 0.16 |
| NADH | 0.19 ± 0.03 | 2.90 ± 0.78 | 5.64 ± 0.23 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mu, X.; Feng, X.; Wu, T.; Zhou, F.; Nie, Y.; Xu, Y. Transamination-Like Reaction Catalyzed by Leucine Dehydrogenase for Efficient Co-Synthesis of α-Amino Acids and α-Keto Acids. Molecules 2021, 26, 7287. https://doi.org/10.3390/molecules26237287
Mu X, Feng X, Wu T, Zhou F, Nie Y, Xu Y. Transamination-Like Reaction Catalyzed by Leucine Dehydrogenase for Efficient Co-Synthesis of α-Amino Acids and α-Keto Acids. Molecules. 2021; 26(23):7287. https://doi.org/10.3390/molecules26237287
Chicago/Turabian StyleMu, Xiaoqing, Xian Feng, Tao Wu, Feng Zhou, Yao Nie, and Yan Xu. 2021. "Transamination-Like Reaction Catalyzed by Leucine Dehydrogenase for Efficient Co-Synthesis of α-Amino Acids and α-Keto Acids" Molecules 26, no. 23: 7287. https://doi.org/10.3390/molecules26237287






