Gas Chromatographic Fingerprint Analysis for the Comparison of Seized Cannabis Samples
Abstract
:1. Introduction
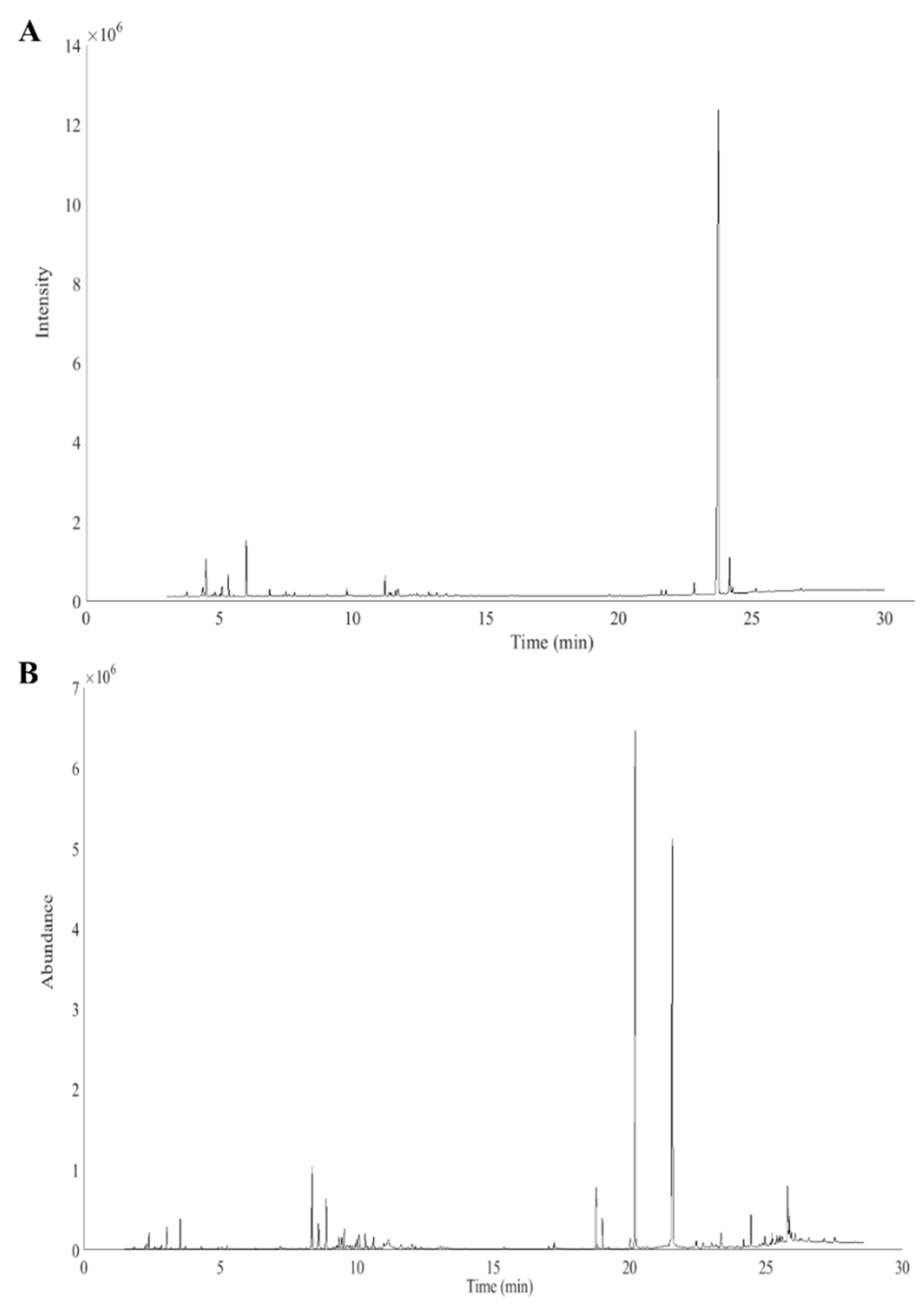
2. Results and Discussion
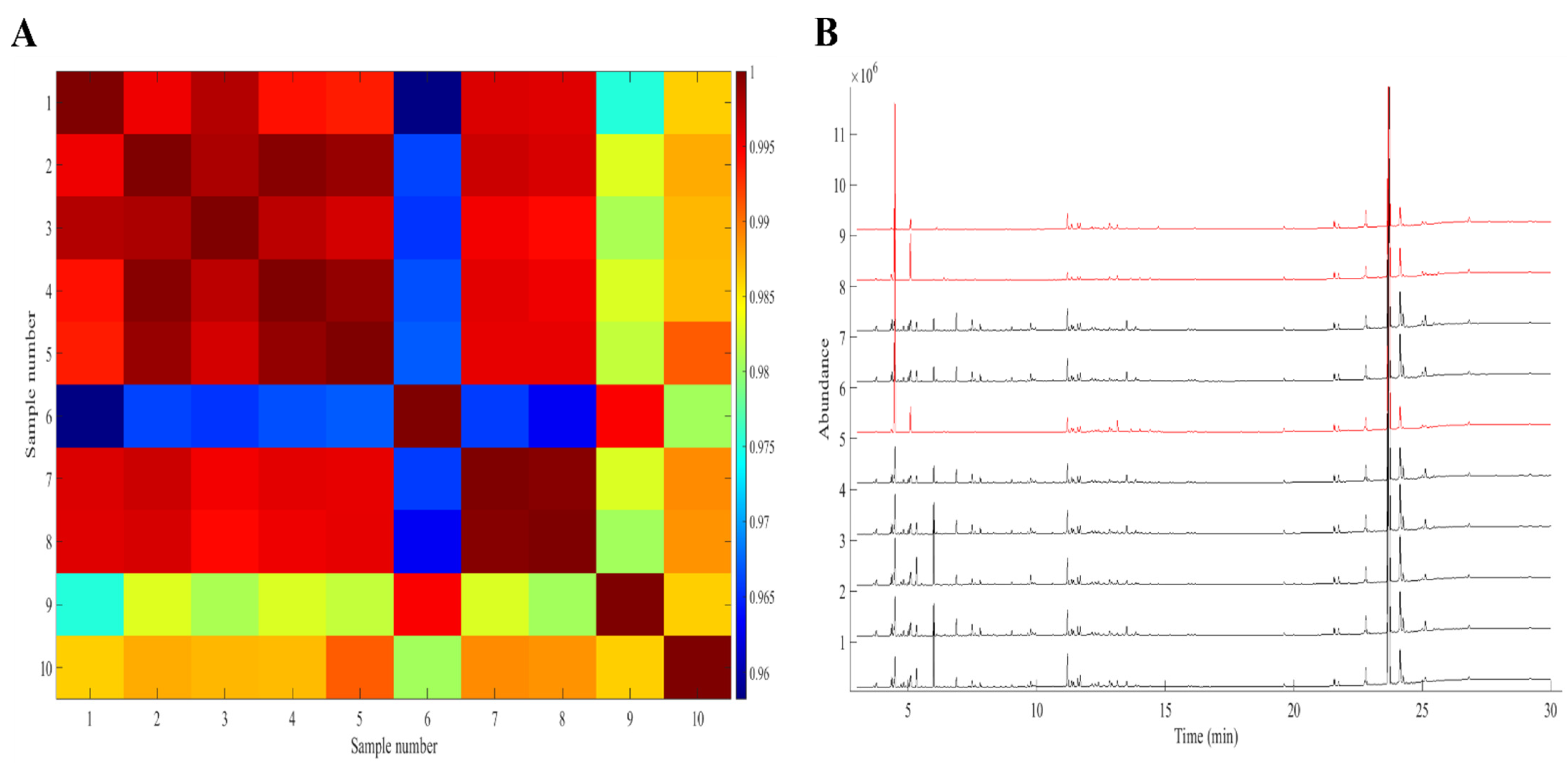
2.1. Similarity of the Cannabis Fingerprints
2.2. Pre-Processing of the Raw Data
2.2.1. Alignment Optimization
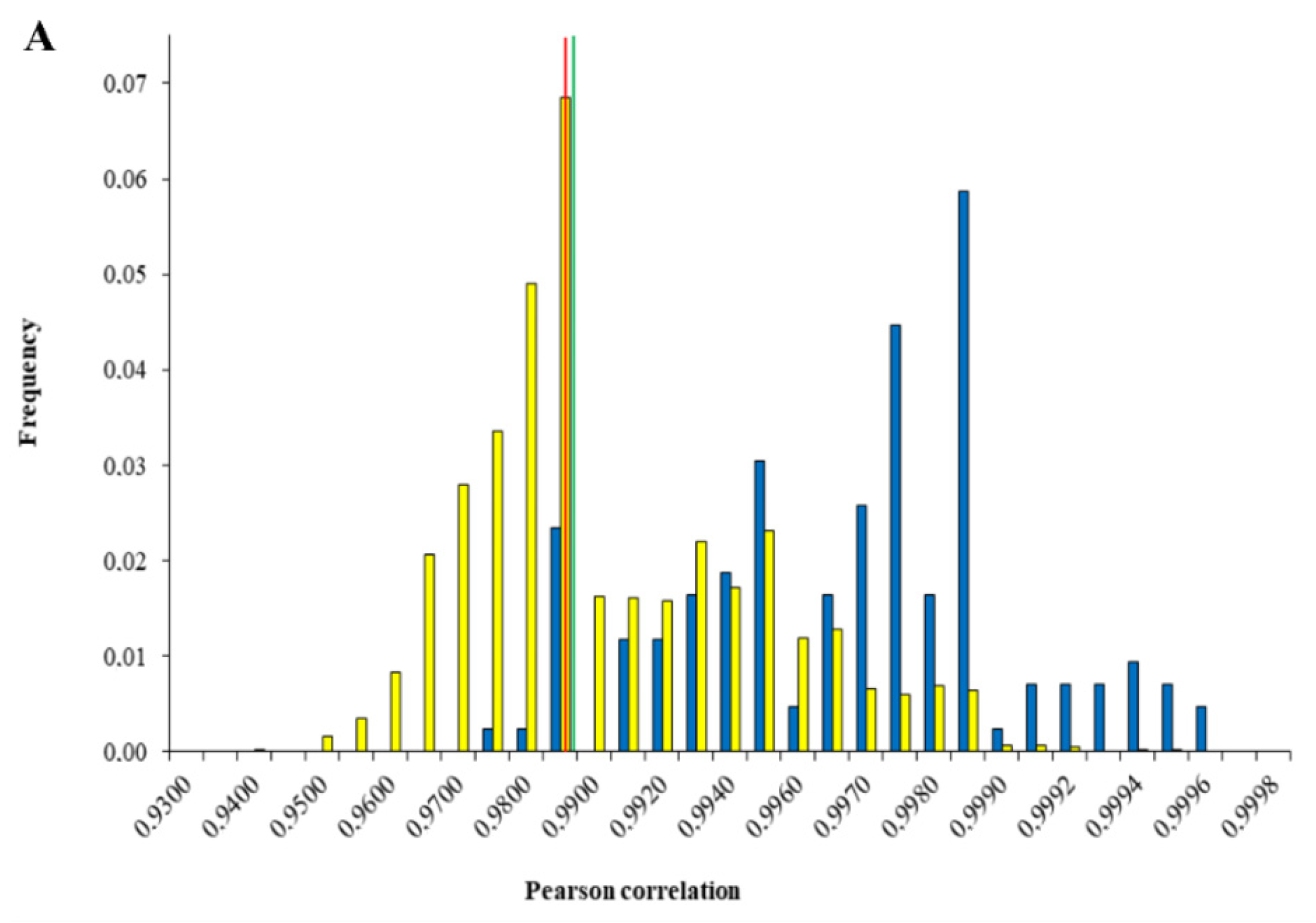
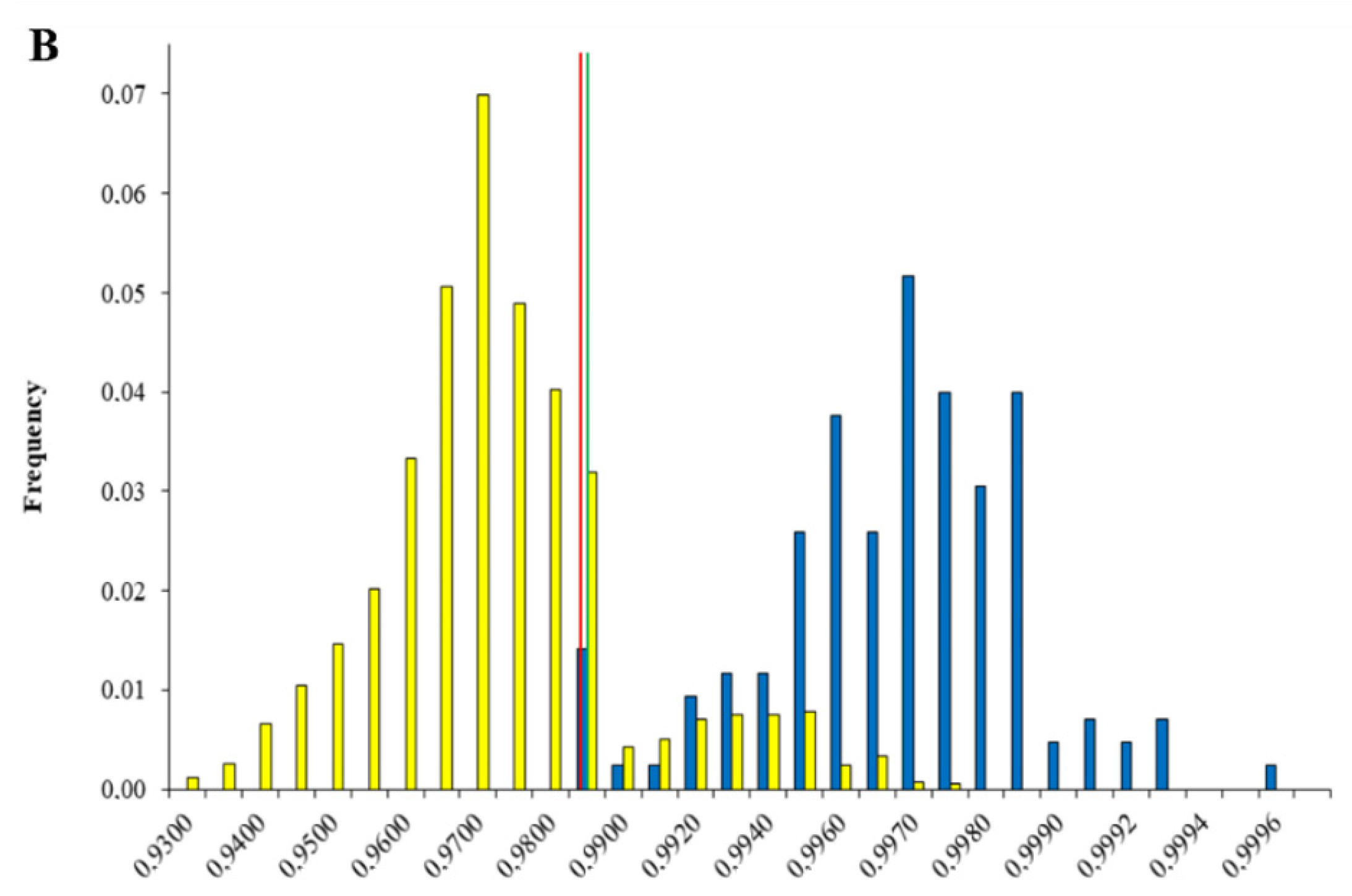
2.2.2. Data Treatment—Discrimination Linked and Unlinked Cannabis Samples
2.3. THC Influence
2.4. Cross-Validation
3. Materials and Methods
3.1. Origin of Samples
3.2. Sample Preparation
3.3. GC–MS Conditions
3.4. GC–FID Conditions
3.5. Data Analysis
3.5.1. Alignment Optimization
Automated Correlation-Optimized Warping (ACOW)
Design of Experiments (DoE)
3.5.2. Data Pre-Processing Techniques
3.5.3. Pearson Correlation Coefficient
3.5.4. False Negative and False Positive Error Rates in Forensic Science
Discriminating Power Methodology
ROC Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Punja, Z.K.; Collyer, D.; Scott, C.; Lung, S.; Holmes, J.; Sutton, D. Pathogens and Molds Affecting Production and Quality of Cannabis sativa L. Front. Plant Sci. 2019, 10, 1120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borille, B.T.; Marcelo, M.C.A.; Ortiz, R.S.; Mariotti, K.D.C.; Ferrão, M.; Limberger, R.P. Near infrared spectroscopy combined with chemometrics for growth stage classification of cannabis cultivated in a greenhouse from seized seeds. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2017, 173, 318–323. [Google Scholar] [CrossRef]
- Cox, C. The Canadian Cannabis Act legalizes and regulates recreational cannabis use in 2018. Health Policy 2018, 122, 205–209. [Google Scholar] [CrossRef] [PubMed]
- Laqueur, H.; Rivera-Aguirre, A.; Shev, A.; Castillo-Carniglia, A.; Rudolph, K.E.; Ramirez, J.; Martins, S.; Cerdá, M. The impact of cannabis legalization in Uruguay on adolescent cannabis use. Int. J. Drug Policy 2020, 80, 102748. [Google Scholar] [CrossRef]
- Sampson, P.B. Phytocannabinoid Pharmacology: Medicinal Properties of Cannabis sativa Constituents Aside from the “Big Two”. J. Nat. Prod. 2021, 84, 142–160. [Google Scholar] [CrossRef] [PubMed]
- Sarris, J.; Sinclair, J.; Karamacoska, D.; Davidson, M.; Firth, J. Medicinal cannabis for psychiatric disorders: A clinically-focused systematic review. BMC Psychiatry 2020, 20, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gracie, K.; Hancox, R.J. Cannabis use disorder and the lungs. Addiction 2021, 116, 182–190. [Google Scholar] [CrossRef]
- European Monitoring Centre for Drugs and Drug Addiction. European Drug Report 2020: Trends and Developments; Publications Office of the European Union: Luxembourg, 2020; Available online: https://data.europa.eu/doi/10.2810/420678 (accessed on 12 April 2021).
- Nic Daeid, N.; Waddell, R.J. The analytical and chemometric procedures used to profile illicit drug seizures. Talanta 2005, 67, 280–285. [Google Scholar] [CrossRef]
- Esseiva, P.; Ioset, S.; Anglada, F.; Gasté, L.; Ribaux, O.; Margot, P.; Gallusser, A.; Biedermann, A.; Specht, Y.; Ottinger, E. Forensic drug Intelligence: An important tool in law enforcement. Forensic Sci. Int. 2007, 167, 247–254. [Google Scholar] [CrossRef]
- Popovic, A.; Morelato, M.; Roux, C.; Beavis, A. Review of the most common chemometric techniques in illicit drug profiling. Forensic Sci. Int. 2019, 302, 109911. [Google Scholar] [CrossRef]
- Rivera, D.; Mozayani, A.; Obi, E.; Drake, J.M. The Structural Complexities of Cannabis sativa L. and Profiling Techniques for Geographic Source Determination. J. Forensic Res. 2019, 10, 1000443. [Google Scholar]
- Lociciro, S.; Esseiva, P.; Hayoz, P.; Dujourdy, L.; Besacier, F.; Margot, P. Cocaine profiling for strategic intelligence, a cross-border project between France and Switzerland: Part II. Validation of the statistical methodology for the profiling of cocaine. Forensic Sci. Int. 2008, 177, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Andersson, K.; Lock, E.; Jalava, K.; Huizer, H.; Jonson, S.; Kaa, E.; Lopes, A.; der Meer, A.P.-V.; Sippola, E.; Dujourdy, L.; et al. Development of a harmonised method for the profiling of amphetamines VI: Evaluation of methods for comparison of amphetamine. Forensic Sci. Int. 2007, 169, 86–99. [Google Scholar] [CrossRef] [PubMed]
- Esseiva, P.; Dujourdy, L.; Anglada, F.; Taroni, F.; Margot, P. A methodology for illicit heroin seizures comparison in a drug intelligence perspective using large databases. Forensic Sci. Int. 2003, 132, 139–152. [Google Scholar] [CrossRef]
- Sexton, M.; Ziskind, J. Sampling Cannabis for Analytical Purposes. 2013. Available online: https://lcb.wa.gov/publications/Marijuana/BOTEC%20reports/1e-Sampling-Lots-Final.pdf (accessed on 11 May 2021).
- Aizpurua-Olaizola, O.; Omar, J.; Navarro, P.; Olivares, M.; Etxebarria, N.; Usobiaga, A. Identification and quantification of cannabinoids in Cannabis sativa L. plants by high performance liquid chromatography-mass spectrometry. Anal. Bioanal. Chem. 2014, 406, 7549–7560. [Google Scholar] [CrossRef]
- ElSohly, M.A. Marijuana and the Cannabinoids; Humana Press: Totowa, NJ, USA, 2007; pp. 17–49. [Google Scholar]
- Hädener, M.; König, S.; Weinmann, W. Quantitative determination of CBD and THC and their acid precursors in confiscated cannabis samples by HPLC-DAD. Forensic Sci. Int. 2019, 299, 142–150. [Google Scholar] [CrossRef] [PubMed]
- ElSohly, M.A.; Chandra, S.; Radwan, M.; Majumdar, C.G.; Church, J.C. A Comprehensive Review of Cannabis Potency in the United States in the Last Decade. Biol. Psychiatry Cogn. Neurosci. Neuroimaging 2021, 6, 603–606. [Google Scholar] [CrossRef]
- Vanhove, W.; Van Damme, P.; Meert, N. Factors determining yield and quality of illicit indoor cannabis (Cannabis spp.) production. Forensic Sci. Int. 2011, 212, 158–163. [Google Scholar] [CrossRef]
- Miró, M.; Worsfold, P.; Poole, C.; Townshend, A. Encyclopedia of Analytical Science, 3rd ed.; Elsevier: Amsterdam, The Netherlands, 2019. [Google Scholar]
- Komsta, Ł.; Vander Heyden, Y.; Sherma, J. (Eds.) Chemometrics in Chromatography; CRC Press: Boca Raton, FL, USA, 2020. [Google Scholar]
- Cieśla, Ł. Biological Fingerprinting of Herbal Samples by Means of Liquid Chromatography. Chromatogr. Res. Int. 2012, 2012, 532418. [Google Scholar] [CrossRef] [Green Version]
- Tistaert, C.; Dejaegher, B.; Heyden, Y.V. Chromatographic separation techniques and data handling methods for herbal fingerprints: A review. Anal. Chim. Acta 2011, 690, 148–161. [Google Scholar] [CrossRef]
- World Health Organization. General Guidelines for Methodologies on Research and Evaluation of Traditional Medicine. 2000. Available online: http://apps.who.int/iris/bitstream/handle/1066/6678/WHO_EDM_TRM_2000.1.pdf;jsessionid=70B972C1A6E0CA1709E1FB0A684CFA5C?sequence=1 (accessed on 25 May 2021).
- Huang, M.-Y.; Zhang, L.-L.; Ding, J.; Lu, J.-J. Anticancer drug discovery from Chinese medicinal herbs. Chin. Med. 2018, 13, 35. [Google Scholar] [CrossRef] [Green Version]
- Fischedick, J.T.; Hazekamp, A.; Erkelens, T.; Choi, Y.H.; Verpoorte, R. Metabolic fingerprinting of Cannabis sativa L., cannabinoids and terpenoids for chemotaxonomic and drug standardization purposes. Phytochemistry 2010, 71, 2058–2073. [Google Scholar] [CrossRef]
- Slosse, A.; Van Durme, F.; Samyn, N.; Mangelings, D.; Heyden, Y.V. Evaluation of data preprocessings for the comparison of GC–MS chemical profiles of seized cannabis samples. Forensic Sci. Int. 2020, 310, 110228. [Google Scholar] [CrossRef]
- Broséus, J.; Huhtala, S.; Esseiva, P. First systematic chemical profiling of cocaine police seizures in Finland in the framework of an intelligence-led approach. Forensic Sci. Int. 2015, 251, 87–94. [Google Scholar] [CrossRef]
- De Backer, B.; Maebe, K.; Verstraete, A.; Charlier, C. Evolution of the Content of THC and Other Major Cannabinoids in Drug-Type Cannabis Cuttings and Seedlings During Growth of Plants. J. Forensic Sci. 2012, 57, 918–922. [Google Scholar] [CrossRef]
- Ben Ahmed, Z.; Yousfi, M.; Viaene, J.; Dejaegher, B.; Demeyer, K.; Mangelings, D.; Heyden, Y.V. Antioxidant activities of Pistacia atlantica extracts modeled as a function of chromatographic fingerprints in order to identify antioxidant markers. Microchem. J. 2016, 128, 208–217. [Google Scholar] [CrossRef]
- Xu, C.-J.; Liang, Y.-Z.; Chau, F.-T.; Heyden, Y.V. Pretreatments of chromatographic fingerprints for quality control of herbal medicines. J. Chromatogr. A 2006, 1134, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Marquis, R.; Weyermann, C.; Delaporte, C.; Esseiva, P.; Aalberg, L.; Besacier, F.; Bozenko, J.S.; Dahlenburg, R.; Kopper, C.; Zrcek, F. Drug intelligence based on MDMA tablets data: 2. Physical characteristics profiling. Forensic Sci. Int. 2008, 178, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; De Souza, D.P.; Keen, W.W.; Saunders, E.C.; McConville, M.J.; Speed, T.P.; A Likić, V. A dynamic programming approach for the alignment of signal peaks in multiple gas chromatography-mass spectrometry experiments. BMC Bioinform. 2007, 8, 419. [Google Scholar] [CrossRef] [Green Version]
- Møller, S.F.; Jorgensen, B.M. Peak alignment and robust principal component analysis of gas chromatograms of fatty acid methyl esters and volatiles. J. Chromatogr. Sci. 2007, 45, 169–176. [Google Scholar] [CrossRef] [Green Version]
- Jiao, L.; Bing, S.; Wang, X.; Xue, Z.; Li, H. The Application of Automated Correlation Optimized Warping to the Quality Evaluation of Radix Puerariae thomsonii: Correcting Retention Time Shift in the Chromatographic Fingerprints. Química Nova 2014, 38, 8–13. [Google Scholar] [CrossRef]
- Tomasi, G.; Berg, F.V.D.; Andersson, C. Correlation optimized warping and dynamic time warping as preprocessing methods for chromatographic data. J. Chemom. 2004, 18, 231–241. [Google Scholar] [CrossRef]
- Skov, T.; Berg, F.V.D.; Tomasi, G.; Bro, R. Automated alignment of chromatographic data. J. Chemom. 2006, 20, 484–497. [Google Scholar] [CrossRef]
- Pravdova, V.; Walczak, B.; Massart, D. A comparison of two algorithms for warping of analytical signals. Anal. Chim. Acta 2002, 456, 77–92. [Google Scholar] [CrossRef]
- Tistaert, C.; Dejaegher, B.; Chataigné, G.; Rivière, C.; Hoai, N.N.; Van, M.C.; Quetin-Leclercq, J.; Heyden, Y.V. Potential antioxidant compounds in Mallotus species fingerprints. Part II: Fingerprint alignment, data analysis and peak identification. Anal. Chim. Acta 2012, 721, 35–43. [Google Scholar] [CrossRef]
- Kessaissia, F.Z.; Zegaoui, A.; Aillerie, M.; Arab, M.; Boutoubat, M.; Fares, C. Factorial design and response surface optimization for modeling photovoltaic module parameters. Energy Rep. 2020, 6, 299–309. [Google Scholar] [CrossRef]
- Tarley, C.R.T.; Silveira, G.; dos Santos, W.N.L.; Matos, G.D.; da Silva, E.G.P.; Bezerra, M.A.; Miró, M.; Ferreira, S.L.C. Chemometric tools in electroanalytical chemistry: Methods for optimization based on factorial design and response surface methodology. Microchem. J. 2009, 92, 58–67. [Google Scholar] [CrossRef]
- Viaene, J.; Heyden, Y.V. Experimental design-based optimization strategies for chromatographic and capillary electrophoretic separations. Handb. Anal. Sep. 2020, 8, 197–275. [Google Scholar] [CrossRef]
- Esseiva, P.; Gaste, L.; Alvarez, D.; Anglada, F. Illicit drug profiling, reflection on statistical comparisons. Forensic Sci. Int. 2011, 207, 27–34. [Google Scholar] [CrossRef]
- Viaene, J.; Goodarzi, M.; Dejaegher, B.; Tistaert, C.; Le Tuan, A.H.; Hoai, N.N.; Van, M.C.; Quetin-Leclercq, J.; Heyden, Y.V. Discrimination and classification techniques applied on Mallotus and Phyllanthus high performance liquid chromatography fingerprints. Anal. Chim. Acta 2015, 877, 41–50. [Google Scholar] [CrossRef]
- Putri, S.P.; Fukusaki, E. Mass Spectrometry-Based Metabolomics a Practical Guide, 1st ed.; CRC Press: Boca Raton, FL, USA, 2016. [Google Scholar]
- Detroyer, A.; Schoonjans, V.; Questier, F.; Heyden, Y.V.; Borosy, A.; Guo, Q.; Massart, D. Exploratory chemometric analysis of the classification of pharmaceutical substances based on chromatographic data. J. Chromatogr. A 2000, 897, 23–36. [Google Scholar] [CrossRef]
- Tistaert, C.; Dejaegher, B.; Hoai, N.N.; Chataigné, G.; Rivière, C.; Hong, V.N.T.; Van, M.C.; Quetin-Leclercq, J.; Heyden, Y.V. Potential antioxidant compounds in Mallotus species fingerprints. Part I: Indication, using linear multivariate calibration techniques. Anal. Chim. Acta 2009, 652, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Segers, K.; Slosse, A.; Viaene, J.; Bannier, M.A.; Van de Kant, K.D.; Dompeling, E.; Van Eeckhaut, A.; Vercammen, J.; Heyden, Y.V. Feasibility study on exhaled-breath analysis by untargeted Selected-Ion Flow-Tube Mass Spectrometry in children with cystic fibrosis, asthma, and healthy controls: Comparison of data pretreatment and classification techniques. Talanta 2021, 225, 122080. [Google Scholar] [CrossRef] [PubMed]
- Akoglu, H. User’s guide to correlation coefficients. Turk. J. Emerg. Med. 2018, 18, 91–93. [Google Scholar] [CrossRef] [PubMed]
- Bell, S. Encyclopedia of Forensic Science, Revised ed.; Facts on File: New York, NY, USA, 2008. [Google Scholar]
- Murrie, D.C.; Gardner, B.O.; Kelley, S.; Dror, I.E. Perceptions and estimates of error rates in forensic science: A survey of forensic analysts. Forensic Sci. Int. 2019, 302, 109887. [Google Scholar] [CrossRef] [Green Version]
- Du, M. Analysis of errors in forensic science. J. Forensic Sci. Med. 2017, 3, 139. [Google Scholar] [CrossRef]
- Park, S.H.; Goo, J.M.; Jo, C.-H. Receiver Operating Characteristic (ROC) Curve: Practical Review for Radiologists. Korean J. Radiol. 2004, 5, 11–18. [Google Scholar] [CrossRef] [Green Version]

| GC–FID | GC–MS | |||||||
|---|---|---|---|---|---|---|---|---|
| Pre-Treatment Method | 95% CL | 99% CL | 95% CL | 99% CL | ||||
| FN (%) | FP (%) | FN (%) | FP (%) | FN (%) | FP (%) | FN (%) | FP (%) | |
| Aligned data (1×ACOW) | 6 | 57 | 4 | 65 | 6 | 54 | 2 | 57 |
| Column centring (CC) | 9 | 55 | 5 | 65 | 7 | 51 | 2 | 56 |
| Normalization and CC (N and CC) | 6 | 57 | 4 | 64 | 5 | 48 | 2 | 53 |
| Standard normal variate and CC (SNV and CC) | 7 | 52 | 4 | 65 | 5 | 48 | 2 | 53 |
| Square root | 6 | 30 | 4 | 39 | 4 | 38 | 2 | 46 |
| Fourth root | 6 | 24 | 4 | 32 | 6 | 28 | 2 | 35 |
| Auto-scaling | 6 | 19 | 4 | 27 | 6 | 64 | 0 | 87 |
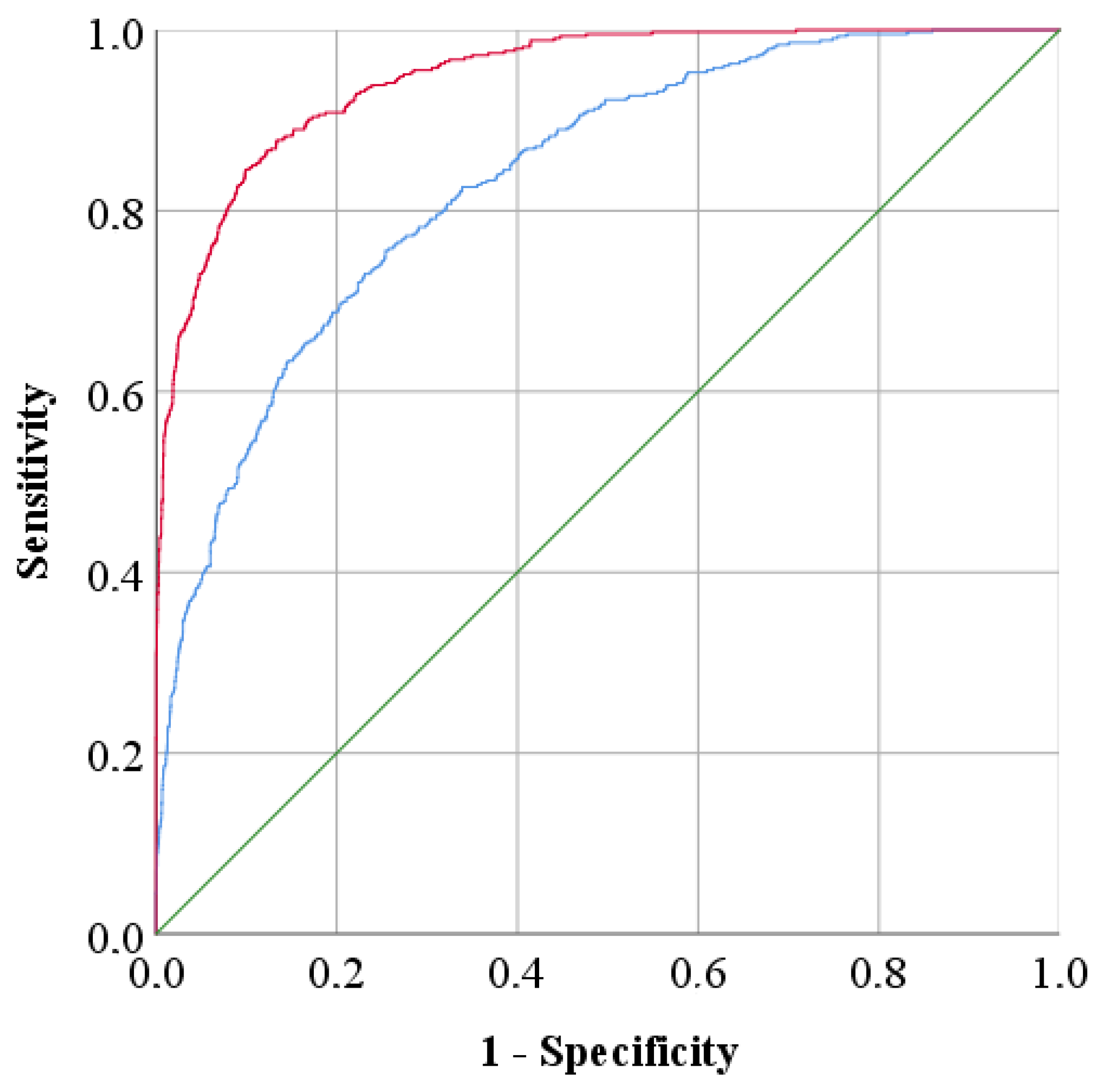
| Data | AUC | 95% Confidence Interval | |
|---|---|---|---|
| Lower Limit | Upper Limit | ||
| Reference | 0.834 | 0.815 | 0.853 |
| After Fourth Root Normalization | 0.947 | 0.938 | 0.957 |
| % Misclassifications | ||||
|---|---|---|---|---|
| Cross-Validation Approach | 95% CI Limit | 99% CI Limit | ||
| FN | FP | FN | FP | |
| Leave-n-Out (LNO) | 6 | 24 | 4 | 32 |
| Leave-One Plantation-Out (LOPO) | 7 | 25 | 4 | 33 |
| Entire Calibration Dataset | 6 | 24 | 4 | 32 |
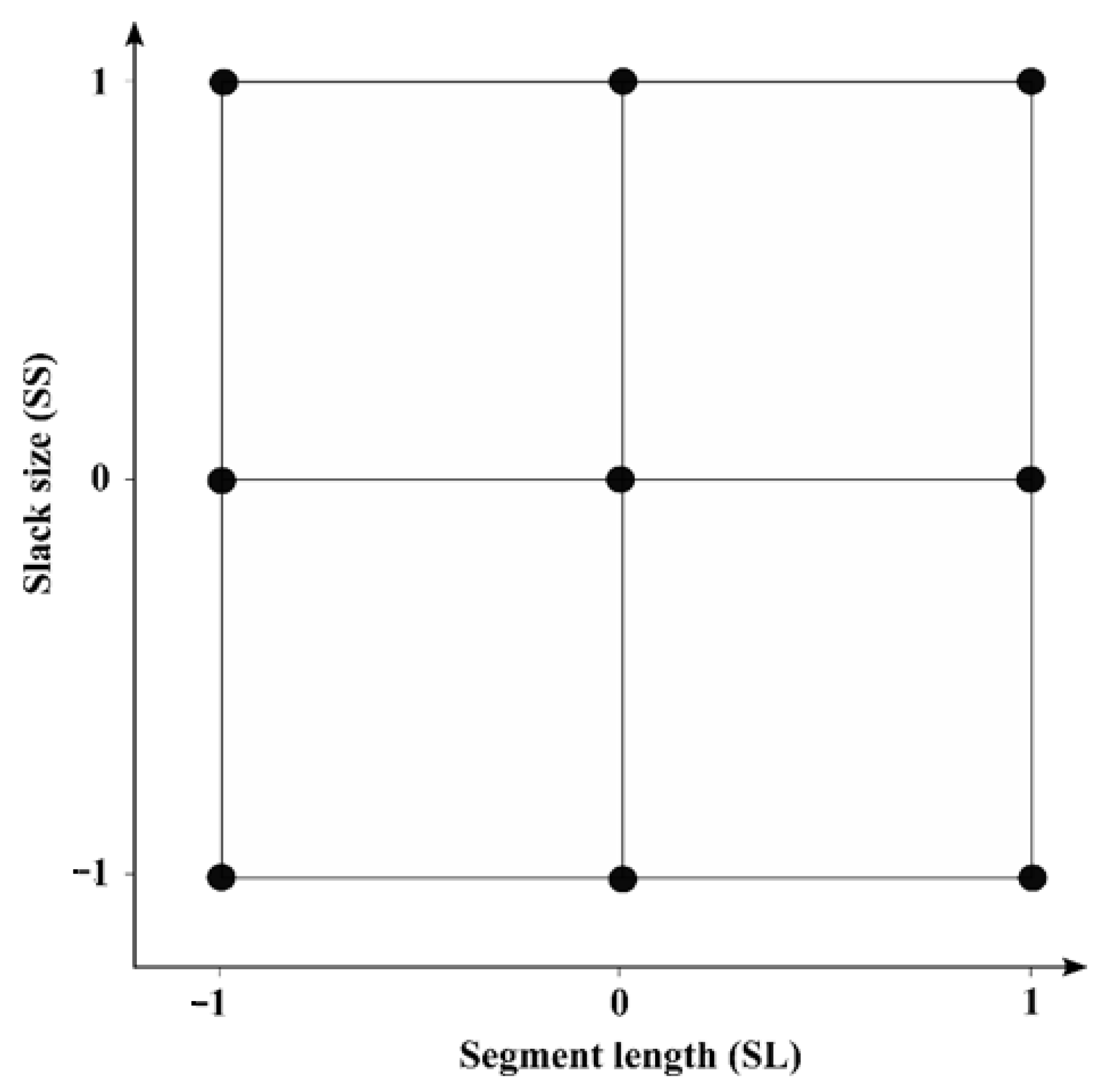
| Design | Segment Length (SL) | Slack Size (SS) | ||||
|---|---|---|---|---|---|---|
| −1 | 0 | 1 | −1 | 0 | 1 | |
| 1 | 15 | 58 | 100 | 1 | 6 | 10 |
| 2 | 25 | 113 | 200 | 1 | 6 | 10 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Slosse, A.; Van Durme, F.; Samyn, N.; Mangelings, D.; Vander Heyden, Y. Gas Chromatographic Fingerprint Analysis for the Comparison of Seized Cannabis Samples. Molecules 2021, 26, 6643. https://doi.org/10.3390/molecules26216643
Slosse A, Van Durme F, Samyn N, Mangelings D, Vander Heyden Y. Gas Chromatographic Fingerprint Analysis for the Comparison of Seized Cannabis Samples. Molecules. 2021; 26(21):6643. https://doi.org/10.3390/molecules26216643
Chicago/Turabian StyleSlosse, Amorn, Filip Van Durme, Nele Samyn, Debby Mangelings, and Yvan Vander Heyden. 2021. "Gas Chromatographic Fingerprint Analysis for the Comparison of Seized Cannabis Samples" Molecules 26, no. 21: 6643. https://doi.org/10.3390/molecules26216643






