Fluorescent and Electron-Dense Green Color Emitting Nanodiamonds for Single-Cell Correlative Microscopy
Abstract
:1. Introduction
2. Results
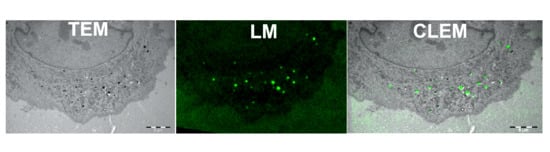
2.1. TEM of Green FNDs in Cells
2.2. LM of Green FNDs
2.3. Correlation between TEM and LM Images
3. Discussion
4. Materials and Methods
4.1. Green Fluorescent Nanodiamonds (Green FNDs)
4.2. Cell Culture
4.3. Transmission Electron Microscopy (TEM)
4.4. Confocal Microscopy
4.5. Image Correlation
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lucas, M.S.; Günthert, M.; Gasser, P.; Lucas, F.; Wepf, R. Bridging Microscopes. In Methods in Cell Biology; Elsevier: Amsterdam, The Netherlands, 2012; Volume 111, pp. 325–356. [Google Scholar]
- Mironov, A.A.; Beznoussenko, G.V. Correlative light-electron microscopy a potent tool for the imaging of rare or unique cellular and tissue events and structures. Methods Enzymol. 2012, 504, 201–219. [Google Scholar] [CrossRef] [PubMed]
- Van Rijnsoever, C.; Oorschot, V.; Klumperman, J. Correlative light-electron microscopy (CLEM) combining live-cell imaging and immunolabeling of ultrathin cryosections. Nat. Methods 2008, 5, 973–980. [Google Scholar] [CrossRef] [PubMed]
- de Boer, P.; Hoogenboom, J.P.; Giepmans, B.N.G. Correlated light and electron microscopy: Ultrastructure lights up! Nat. Methods 2015, 12, 503–513. [Google Scholar] [CrossRef] [PubMed]
- Polishchuk, E.V.; Polishchuk, R.S. Pre-embedding labeling for subcellular detection of molecules with electron microscopy. Tissue Cell 2018, 57, 103–110. [Google Scholar] [CrossRef]
- Ando, T.; Bhamidimarri, S.P.; Brending, N.; Colin-York, H.; Collinson, L.; De Jonge, N.; De Pablo, P.J.; Debroye, E.; Eggeling, C.; Franck, C.; et al. The 2018 correlative microscopy techniques roadmap. J. Phys. D. Appl. Phys. 2018, 51, 443001. [Google Scholar] [CrossRef] [Green Version]
- Loussert Fonta, C.; Humbel, B.M. Correlative microscopy. Arch. Biochem. Biophys. 2015, 581, 98–110. [Google Scholar] [CrossRef]
- Kopek, B.G.; Shtengel, G.; Xu, C.S.; Clayton, D.A.; Hess, H.F. Correlative 3D superresolution fluorescence and electron microscopy reveal the relationship of mitochondrial nucleoids to membranes. Proc. Natl. Acad. Sci. USA 2012, 109, 6136–6141. [Google Scholar] [CrossRef] [Green Version]
- Ganeva, I.; Kukulski, W. Membrane Architecture in the Spotlight of Correlative Microscopy. Trends Cell Biol. 2020, 30, 577–587. [Google Scholar] [CrossRef]
- Polishchuk, R.S.; Polishchuk, E.V.; Luini, A. Chapter 2—Visualizing Live Dynamics and Ultrastructure of Intracellular Organelles with Preembedding Correlative Light-Electron Microscopy. In Methods in Cell Biology; Verkade, T.M.-R., Paul, V., Eds.; Correlative Light and Electron {MIcroscopy}; Academic Press: Cambridge, MA, USA, 2012; Volume 111, pp. 21–35. [Google Scholar]
- Kukulski, W.; Schorb, M.; Welsch, S.; Picco, A.; Kaksonen, M.; Briggs, J.A.G. Correlated fluorescence and 3D electron microscopy with high sensitivity and spatial precision. J. Cell Biol. 2011, 192, 111–119. [Google Scholar] [CrossRef]
- Booth, D.G.; Beckett, A.J.; Molina, O.; Samejima, I.; Masumoto, H.; Kouprina, N.; Larionov, V.; Prior, I.A.; Earnshaw, W.C. 3D-CLEM Reveals that a Major Portion of Mitotic Chromosomes Is Not Chromatin. Mol. Cell 2016, 64, 790–802. [Google Scholar] [CrossRef]
- Biazik, J.; Vihinen, H.; Anwar, T.; Jokitalo, E.; Eskelinen, E.-L. The versatile electron microscope: An ultrastructural overview of autophagy. Methods 2015, 75, 44–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abbe, E. Beiträge zur Theorie des Mikroskops und der mikroskopischen Wahrnehmung: I. Die Construction von Mikroskopen auf Grund der Theorie. Arch. Für Mikrosk. Anat. 1873, 9, 413–418. [Google Scholar] [CrossRef]
- Klar, T.A.; Jakobs, S.; Dyba, M.; Egner, A.; Hell, S.W. Fluorescence microscopy with diffraction resolution barrier broken by stimulated emission. Proc. Natl. Acad. Sci. USA 2000, 97, 8206–8210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hell, S.W.; Wichmann, J. stimulated-emission-depletion fluorescence microscopy. Opt. Lett. 1994, 19, 780–782. [Google Scholar] [CrossRef] [PubMed]
- Wolff, G.; Hagen, C.; Grünewald, K.; Kaufmann, R. Towards correlative super-resolution fluorescence and electron cryo-microscopy. Biol. Cell 2016, 108, 245–258. [Google Scholar] [CrossRef] [Green Version]
- Johnson, E.; Seiradake, E.; Jones, E.Y.; Davis, I.; Grünewald, K.; Kaufmann, R. Correlative in-resin super-resolution and electron microscopy using standard fluorescent proteins. Sci. Rep. 2015, 5, 9583. [Google Scholar] [CrossRef]
- Prabhakar, N.; Peurla, M.; Koho, S.; Deguchi, T.; Näreoja, T.; Chang, H.-C.; Rosenholm, J.M.; Hänninen, P.E. STED-TEM Correlative Microscopy Leveraging Nanodiamonds as Intracellular Dual-Contrast Markers. Small 2018, 14, 1701807. [Google Scholar] [CrossRef]
- Watanabe, S.; Punge, A.; Hollopeter, G.; Willig, K.I.; Hobson, R.J.; Davis, M.W.; Hell, S.W.; Jorgensen, E.M. Protein localization in electron micrographs using fluorescence nanoscopy. Nat. Methods 2011, 8, 80–84. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Deerinck, T.J.; Sigal, Y.M.; Babcock, H.P.; Ellisman, M.H.; Zhuang, X. Correlative Stochastic Optical Reconstruction Microscopy and Electron Microscopy. PLoS ONE 2015, 10, e0124581. [Google Scholar] [CrossRef]
- Tuijtel, M.W.; Koster, A.J.; Jakobs, S.; Faas, F.G.A.; Sharp, T.H. Correlative cryo super-resolution light and electron microscopy on mammalian cells using fluorescent proteins. Sci. Rep. 2019, 9, 1369. [Google Scholar] [CrossRef]
- Paez-Segala, M.G.; Sun, M.G.; Shtengel, G.; Viswanathan, S.; Baird, M.A.; Macklin, J.J.; Patel, R.; Allen, J.R.; Howe, E.S.; Piszczek, G.; et al. Fixation-resistant photoactivatable fluorescent proteins for CLEM. Nat. Methods 2015, 12, 215–218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peddie, C.J.; Domart, M.-C.; Snetkov, X.; O’Toole, P.; Larijani, B.; Way, M.; Cox, S.; Collinson, L.M. Correlative super-resolution fluorescence and electron microscopy using conventional fluorescent proteins in vacuo. J. Struct. Biol. 2017, 199, 120–131. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.-W.; Chen, S.; Tocheva, E.I.; Treuner-Lange, A.; Löbach, S.; Søgaard-Andersen, L.; Jensen, G.J. Correlated cryogenic photoactivated localization microscopy and cryo-electron tomography. Nat. Methods 2014, 11, 737–739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peddie, C.J.; Collinson, L.M. Exploring the third dimension: Volume electron microscopy comes of age. Micron 2014, 61, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Russell, M.R.G.; Lerner, T.R.; Burden, J.J.; Nkwe, D.O.; Pelchen-Matthews, A.; Domart, M.-C.; Durgan, J.; Weston, A.; Jones, M.L.; Peddie, C.J.; et al. 3D correlative light and electron microscopy of cultured cells using serial blockface scanning electron microscopy. J. Cell Sci. 2017, 130, 278–291. [Google Scholar] [CrossRef] [Green Version]
- Sartori, A.; Gatz, R.; Beck, F.; Rigort, A.; Baumeister, W.; Plitzko, J.M. Correlative microscopy: Bridging the gap between fluorescence light microscopy and cryo-electron tomography. J. Struct. Biol. 2007, 160, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Bykov, Y.S.; Cohen, N.; Gabrielli, N.; Manenschijn, H.; Welsch, S.; Chlanda, P.; Kukulski, W.; Patil, K.R.; Schuldiner, M.; Briggs, J.A.G. High-throughput ultrastructure screening using electron microscopy and fluorescent barcoding. J. Cell Biol. 2019, 218, 2797–2811. [Google Scholar] [CrossRef] [Green Version]
- Horstmann, H.; Körber, C.; Sätzler, K.; Aydin, D.; Kuner, T. Serial section scanning electron microscopy (S3EM) on silicon wafers for ultra-structural volume imaging of cells and tissues. PLoS ONE 2012, 7, e35172. [Google Scholar] [CrossRef] [Green Version]
- Knott, G.; Marchman, H.; Wall, D.; Lich, B. Serial section scanning electron microscopy of adult brain tissue using focused ion beam milling. J. Neurosci. 2008, 28, 2959–2964. [Google Scholar] [CrossRef]
- Desbois, G.; Urai, J.L.; Burkhardt, C.; Drury, M.R.; Hayles, M.; Humbel, B. Cryogenic vitrification and 3D serial sectioning using high resolution cryo-FIB SEM technology for brine-filled grain boundaries in halite: First results. Geofluids 2008, 8, 60–72. [Google Scholar] [CrossRef]
- Vihinen, H.; Belevich, I.; Jokitalo, E. Three Dimensional Electron Microscopy of Cellular Organelles by Serial Block Face SEM and ET; Wiley: Hoboken, NJ, USA, 2013; Volume 27. [Google Scholar]
- Kukulski, W.; Schorb, M.; Welsch, S.; Picco, A.; Kaksonen, M.; Briggs, J.A.G. Precise, Correlated Fluorescence Microscopy and Electron Tomography of Lowicryl Sections Using Fluorescent Fiducial Markers. In Methods in Cell Biology; Elsevier: Amsterdam, The Netherlands, 2012; Volume 111, pp. 235–257. [Google Scholar]
- Agronskaia, A.V.; Valentijn, J.A.; van Driel, L.F.; Schneijdenberg, C.T.W.M.; Humbel, B.M.; van Bergen en Henegouwen, P.M.P.; Verkleij, A.J.; Koster, A.J.; Gerritsen, H.C. Integrated fluorescence and transmission electron microscopy. J. Struct. Biol. 2008, 164, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Mohammadian, S.; Fokkema, J.; Agronskaia, A.V.; Liv, N.; de Heus, C.; van Donselaar, E.; Blab, G.A.; Klumperman, J.; Gerritsen, H.C. High accuracy, fiducial marker-based image registration of correlative microscopy images. Sci. Rep. 2019, 9, 3211. [Google Scholar] [CrossRef] [PubMed]
- Nisman, R.; Dellaire, G.; Ren, Y.; Li, R.; Bazett-Jones, D.P. Application of Quantum Dots as Probes for Correlative Fluorescence, Conventional, and Energy-filtered Transmission Electron Microscopy. J. Histochem. Cytochem. 2004, 52, 13–18. [Google Scholar] [CrossRef] [Green Version]
- Deerinck, T.J. The application of fluorescent quantum dots to confocal, multiphoton, and electron microscopic imaging. Toxicol. Pathol. 2008, 36, 112–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Hest, J.J.H.A.; Agronskaia, A.; Fokkema, J.; Montanarella, F.; Puig, A.G.; Donega, C.D.M.; Meijerink, A.; Blab, G.; Gerritsen, H. Towards robust and versatile single nanoparticle fiducial markers for correlative light and electron microscopy. J. Microsc. 2019, 274, 13–22. [Google Scholar] [CrossRef] [Green Version]
- Resch-Genger, U.; Grabolle, M.; Cavaliere-Jaricot, S.; Nitschke, R.; Nann, T. Quantum dots versus organic dyes as fluorescent labels. Nat. Methods 2008, 5, 763–775. [Google Scholar] [CrossRef]
- Chan, W.C.; Nie, S.; Gin, P.; Weiss, S.; Alivisatos, A.P.; Libchaber, A. Quantum Dot Bioconjugates for Ultrasensitive Nonisotopic Detection. Science 1998, 281, 2016–2018. [Google Scholar] [CrossRef] [Green Version]
- Niitsuma, J.; Oikawa, H.; Kimura, E.; Ushiki, T.; Sekiguchi, T. Cathodoluminescence investigation of organic materials. Microscopy 2005, 54, 325–330. [Google Scholar] [CrossRef]
- Rodriguez-Viejo, J.; Jensen, K.F.; Mattoussi, H.; Michel, J.; Dabbousi, B.O.; Bawendi, M.G. Cathodoluminescence and photoluminescence of highly luminescent CdSe/ZnS quantum dot composites. Appl. Phys. Lett. 1997, 70, 2132–2134. [Google Scholar] [CrossRef] [Green Version]
- Hardman, R. A toxicologic review of quantum dots: Toxicity depends on physicochemical and environmental factors. Environ. Health Perspect. 2006, 114, 165–172. [Google Scholar] [CrossRef]
- Tsoi, K.M.; Dai, Q.; Alman, B.A.; Chan, W.C.W. Are Quantum Dots Toxic? Exploring the Discrepancy Between Cell Culture and Animal Studies. Acc. Chem. Res. 2013, 46, 662–671. [Google Scholar] [CrossRef] [PubMed]
- Valizadeh, A.; Mikaeili, H.; Samiei, M.; Farkhani, S.; Zarghami, N.; kouhi, M.; Akbarzadeh, A.; Davaran, S.; Gregory, J.; Salamo, D.; et al. Quantum dots: Synthesis, bioapplications, and toxicity. Nanoscale Res. Lett. 2012, 7, 480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prabhakar, N.; Khan, M.H.; Peurla, M.; Chang, H.-C.; Hänninen, P.E.; Rosenholm, J.M. Intracellular Trafficking of Fluorescent Nanodiamonds and Regulation of Their Cellular Toxicity. ACS Omega 2017, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iakoubovskii, K.; Mitsuishi, K.; Furuya, K. High-resolution electron microscopy of detonation nanodiamond. Nanotechnology 2008, 19, 155705. [Google Scholar] [CrossRef] [PubMed]
- Vlasov, I.I.; Shiryaev, A.A.; Rendler, T.; Steinert, S.; Lee, S.-Y.; Antonov, D.; Vörös, M.; Jelezko, F.; Fisenko, A.V.; Semjonova, L.F.; et al. Molecular-sized fluorescent nanodiamonds. Nat. Nanotechnol. 2014, 9, 54–58. [Google Scholar] [CrossRef] [PubMed]
- Arroyo-Camejo, S.; Adam, M.-P.; Besbes, M.; Hugonin, J.-P.; Jacques, V.; Greffet, J.-J.; Roch, J.-F.; Hell, S.W.; Treussart, F. Stimulated Emission Depletion Microscopy Resolves Individual Nitrogen Vacancy Centers in Diamond Nanocrystals. ACS Nano 2013, 7, 10912–10919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hemelaar, S.R.; de Boer, P.; Chipaux, M.; Zuidema, W.; Hamoh, T.; Martinez, F.P.; Nagl, A.; Hoogenboom, J.P.; Giepmans, B.N.G.; Schirhagl, R.; et al. Nanodiamonds as multi-purpose labels for microscopy. Sci. Rep. 2017, 7, 720. [Google Scholar] [CrossRef]
- Hsieh, F.-J.; Chen, Y.-W.; Huang, Y.-K.; Lee, H.-M.; Lin, C.-H.; Chang, H.-C. Correlative Light-Electron Microscopy of Lipid-Encapsulated Fluorescent Nanodiamonds for Nanometric Localization of Cell Surface Antigens. Anal. Chem. 2018, 90, 1566–1571. [Google Scholar] [CrossRef]
- Liu, W.; Naydenov, B.; Chakrabortty, S.; Wuensch, B.; Hübner, K.; Ritz, S.; Cölfen, H.; Barth, H.; Koynov, K.; Qi, H.; et al. Fluorescent Nanodiamond–Gold Hybrid Particles for Multimodal Optical and Electron Microscopy Cellular Imaging. Nano Lett. 2016, 16, 6236–6244. [Google Scholar] [CrossRef]
- Schrand, A.M.; Hens, S.A.C.; Shenderova, O.A. Nanodiamond Particles: Properties and Perspectives for Bioapplications. Crit. Rev. Solid State Mater. Sci. 2009, 34, 18–74. [Google Scholar] [CrossRef]
- Claveau, S.; Bertrand, J.-R.; Treussart, F.; Claveau, S.; Bertrand, J.-R.; Treussart, F. Fluorescent Nanodiamond Applications for Cellular Process Sensing and Cell Tracking. Micromachines 2018, 9, 247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reineck, P.; Capelli, M.; Lau, D.W.M.; Jeske, J.; Field, M.R.; Ohshima, T.; Greentree, A.D.; Gibson, B.C.; Sreenivasan, V.K.A.; Brown, L.J.; et al. Bright and photostable nitrogen-vacancy fluorescence from unprocessed detonation nanodiamond. Nanoscale 2017, 9, 497–502. [Google Scholar] [CrossRef] [PubMed]
- Tran, T.T.; Kianinia, M.; Bray, K.; Kim, S.; Xu, Z.-Q.; Gentle, A.; Sontheimer, B.; Bradac, C.; Aharonovich, I. Nanodiamonds with photostable, sub-gigahertz linewidth quantum emitters. APL Photonics 2017, 2, 116103. [Google Scholar] [CrossRef] [Green Version]
- Say, J.M.; van Vreden, C.; Reilly, D.J.; Brown, L.J.; Rabeau, J.R.; King, N.J.C. Luminescent nanodiamonds for biomedical applications. Biophys. Rev. 2011, 3, 171–184. [Google Scholar] [CrossRef] [Green Version]
- Davies, G.; Hamer, M.F. Optical Studies of the 1.945 eV Vibronic Band in Diamond. Proc. R. Soc. A Math. Phys. Eng. Sci. 1976, 348, 285–298. [Google Scholar] [CrossRef]
- Treussart, F.; Jacques, V.; Wu, E.; Gacoin, T.; Grangier, P.; Roch, J.-F. Photoluminescence of single colour defects in 50 nm diamond nanocrystals. Phys. B Condens. Matter 2006, 376–377, 926–929. [Google Scholar] [CrossRef] [Green Version]
- Xiao, J.; Liu, P.; Li, L.; Yang, G. Fluorescence Origin of Nanodiamonds. J. Phys. Chem. C 2015, 119, 2239–2248. [Google Scholar] [CrossRef]
- Laporte, G.; Psaltis, D. STED imaging of green fluorescent nanodiamonds containing nitrogen-vacancy-nitrogen centers. Biomed. Opt. Express 2015, 7, 34. [Google Scholar] [CrossRef] [Green Version]
- Prabhakar, N.; Näreoja, T.; Von Haartman, E.; Karaman, D.Ş.; Jiang, H.; Koho, S.; Dolenko, T.A.; Hänninen, P.E.; Vlasov, D.I.; Ralchenko, V.G.; et al. Core-shell designs of photoluminescent nanodiamonds with porous silica coatings for bioimaging and drug delivery II: Application. Nanoscale 2013, 5. [Google Scholar] [CrossRef]
- Wee, T.L.; Tzeng, Y.K.; Han, C.C.; Chang, H.C.; Fann, W.; Hsu, J.H.; Chen, K.M.; Yu, E.C. Two-photon excited fluorescence of nitrogen-vacancy centers in proton-irradiated type Ib diamond. J. Phys. Chem. A 2007, 111, 9379–9386. [Google Scholar] [CrossRef]
- Jimenez, C.M.; Knezevic, N.Z.; Rubio, Y.G.; Szunerits, S.; Boukherroub, R.; Teodorescu, F.; Croissant, J.G.; Hocine, O.; Seric, M.; Raehm, L.; et al. Nanodiamond–PMO for two-photon PDT and drug delivery. J. Mater. Chem. B 2016, 4, 5803–5808. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Fang, C.-Y.; Chang, C.-C.; Peterson, R.; Maswadi, S.; Glickman, R.D.; Chang, H.-C.; Ye, J.Y. Photoacoustic emission from fluorescent nanodiamonds enhanced with gold nanoparticles. Biomed. Opt. Express 2012, 3, 1662–1669. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Cui, H.; Fang, C.-Y.; Jo, J.; Yang, X.; Chang, H.-C.; Forrest, M.L. In vivo photoacoustic imaging of breast cancer tumor with HER2-targeted nanodiamonds. Proc. SPIE Int. Soc. Opt. Eng. 2013, 8815. [Google Scholar] [CrossRef] [Green Version]
- Nunn, N.; Prabhakar, N.; Reineck, P.; Magidson, V.; Kamiya, E.; Heinz, W.F.; Torelli, M.D.; Rosenholm, J.; Zaitsev, A.; Shenderova, O. Brilliant blue, green, yellow, and red fluorescent diamond particles: Synthesis, characterization, and multiplex imaging demonstrations. Nanoscale 2019, 11, 11584–11595. [Google Scholar] [CrossRef]
- Wee, T.-L.; Mau, Y.-W.; Fang, C.-Y.; Hsu, H.-L.; Han, C.-C.; Chang, H.-C. Preparation and characterization of green fluorescent nanodiamonds for biological applications. Diam. Relat. Mater. 2009, 18, 567–573. [Google Scholar] [CrossRef]
- Kang, R.-H.; Baek, S.W.; Ryu, T.-K.; Choi, S.-W. Fabrication of blue-fluorescent nanodiamonds modified with alkyl isocyanate for cellular bioimaging. Colloids Surf. B Biointerfaces 2018, 167, 191–196. [Google Scholar] [CrossRef]
- Liu, K.-K.; Cheng, C.-L.; Chang, C.-C.; Chao, J.-I. Biocompatible and detectable carboxylated nanodiamond on human cell. Nanotechnology 2007, 18, 325102. [Google Scholar] [CrossRef]
- Vaijayanthimala, V.; Chang, H.-C. Functionalized fluorescent nanodiamonds for biomedical applications. Nanomedicine (Lond.) 2009, 4, 47–55. [Google Scholar] [CrossRef]
- Fang, C.-Y.; Vaijayanthimala, V.; Cheng, C.-A.; Yeh, S.-H.; Chang, C.-F.; Li, C.-L.; Chang, H.-C. The Exocytosis of Fluorescent Nanodiamond and Its Use as a Long-Term Cell Tracker. Small 2011, 7, 3363–3370. [Google Scholar] [CrossRef]
- Turcheniuk, K.; Mochalin, V.N. Biomedical applications of nanodiamond (Review). Nanotechnology 2017, 28, 252001. [Google Scholar] [CrossRef]
- Alhaddad, A.; Adam, M.P.; Botsoa, J.; Dantelle, G.; Perruchas, S.; Gacoin, T.; Mansuy, C.; Lavielle, S.; Malvy, C.; Treussart, F.; et al. Nanodiamond as a vector for siRNA delivery to Ewing sarcoma cells. Small 2011, 7, 3087–3095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shenderova, O.A.; McGuire, G.E. Science and engineering of nanodiamond particle surfaces for biological applications (Review). Biointerphases 2015, 10, 030802. [Google Scholar] [CrossRef] [PubMed]
- Bradac, C.; Say, J.M.; Rastogi, I.D.; Cordina, N.M.; Volz, T.; Brown, L.J. Nano-assembly of nanodiamonds by conjugation to actin filaments. J. Biophotonics 2016, 9, 296–304. [Google Scholar] [CrossRef] [PubMed]
- Mkandawire, M.; Pohl, A.; Gubarevich, T.; Lapina, V.; Appelhans, D.; Rödel, G.; Pompe, W.; Schreiber, J.; Opitz, J. Selective targeting of green fluorescent nanodiamond conjugates to mitochondria in HeLa cells. J. Biophotonics 2009, 2, 596–606. [Google Scholar] [CrossRef]
- Zhang, W.; Patel, K.; Schexnider, A.; Banu, S.; Radadia, A.D. Nanostructuring of biosensing electrodes with nanodiamonds for antibody immobilization. ACS Nano 2014, 8, 1419–1428. [Google Scholar] [CrossRef]
- Hui, Y.Y.; Cheng, C.-L.; Chang, H.-C. Nanodiamonds for optical bioimaging. J. Phys. D. Appl. Phys. 2010, 43, 374021. [Google Scholar] [CrossRef]
- Sotoma, S.; Hsieh, F.-J.; Chang, H.-C. Biohybrid fluorescent nanodiamonds as dual-contrast markers for light and electron microscopies. J. Chin. Chem. Soc. 2018, 65, 1136–1146. [Google Scholar] [CrossRef]
- Han, S.; Raabe, M.; Hodgson, L.; Mantell, J.; Verkade, P.; Lasser, T.; Landfester, K.; Weil, T.; Lieberwirth, I. High-Contrast Imaging of Nanodiamonds in Cells by Energy Filtered and Correlative Light-Electron Microscopy: Toward a Quantitative Nanoparticle-Cell Analysis. Nano Lett. 2019, 19, 2178–2185. [Google Scholar] [CrossRef] [Green Version]
- Prabhakar, N.; Belevich, I.; Peurla, M.; Heiligenstein, X.; Chang, H.C.; Sahlgren, C.; Jokitalo, E.; Rosenholm, J.M. Dynamic-ultrastructural cell volume (3D) correlative microscopy facilitated by intracellular fluorescent nanodiamonds as multi-modal probes. bioRxiv 2019. [Google Scholar] [CrossRef] [Green Version]
- Paul-Gilloteaux, P.; Heiligenstein, X.; Belle, M.; Domart, M.-C.; Larijani, B.; Collinson, L.; Raposo, G.; Salamero, J. eC-CLEM: Flexible multidimensional registration software for correlative microscopies. Nat. Methods 2017, 14, 102–103. [Google Scholar] [CrossRef]
- Dei Cas, L.; Zeldin, S.; Nunn, N.; Torelli, M.; Shames, A.I.; Zaitsev, A.M.; Shenderova, O. Fluorescent Diamond Particles: From Fancy Blue to Red: Controlled Production of a Vibrant Color Spectrum of Fluorescent Diamond Particles (Adv. Funct. Mater. 19/2019). Adv. Funct. Mater. 2019, 29, 1970128. [Google Scholar] [CrossRef] [Green Version]
Sample Availability: Not available. |



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prabhakar, N.; Peurla, M.; Shenderova, O.; Rosenholm, J.M. Fluorescent and Electron-Dense Green Color Emitting Nanodiamonds for Single-Cell Correlative Microscopy. Molecules 2020, 25, 5897. https://doi.org/10.3390/molecules25245897
Prabhakar N, Peurla M, Shenderova O, Rosenholm JM. Fluorescent and Electron-Dense Green Color Emitting Nanodiamonds for Single-Cell Correlative Microscopy. Molecules. 2020; 25(24):5897. https://doi.org/10.3390/molecules25245897
Chicago/Turabian StylePrabhakar, Neeraj, Markus Peurla, Olga Shenderova, and Jessica M. Rosenholm. 2020. "Fluorescent and Electron-Dense Green Color Emitting Nanodiamonds for Single-Cell Correlative Microscopy" Molecules 25, no. 24: 5897. https://doi.org/10.3390/molecules25245897