The Function and Mechanism of Lipid Molecules and Their Roles in The Diagnosis and Prognosis of Breast Cancer
Abstract
:1. Introduction
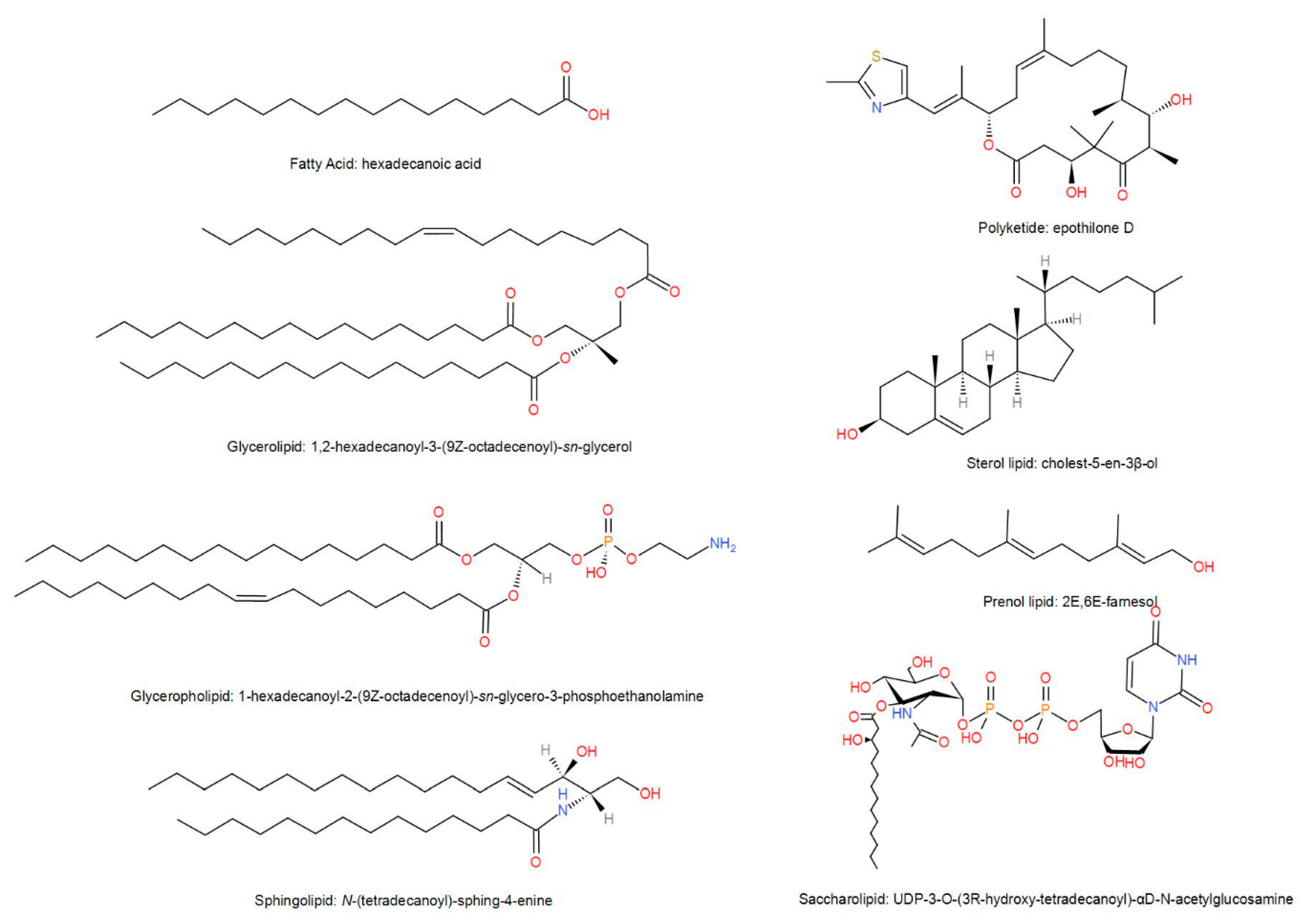
2. Biological Characteristics of Lipids
3. Research Progress of Lipidomics
4. Lipids and Drug Resistance in Cancers
5. Role of Lipid in the Diagnosis of Breast Cancer
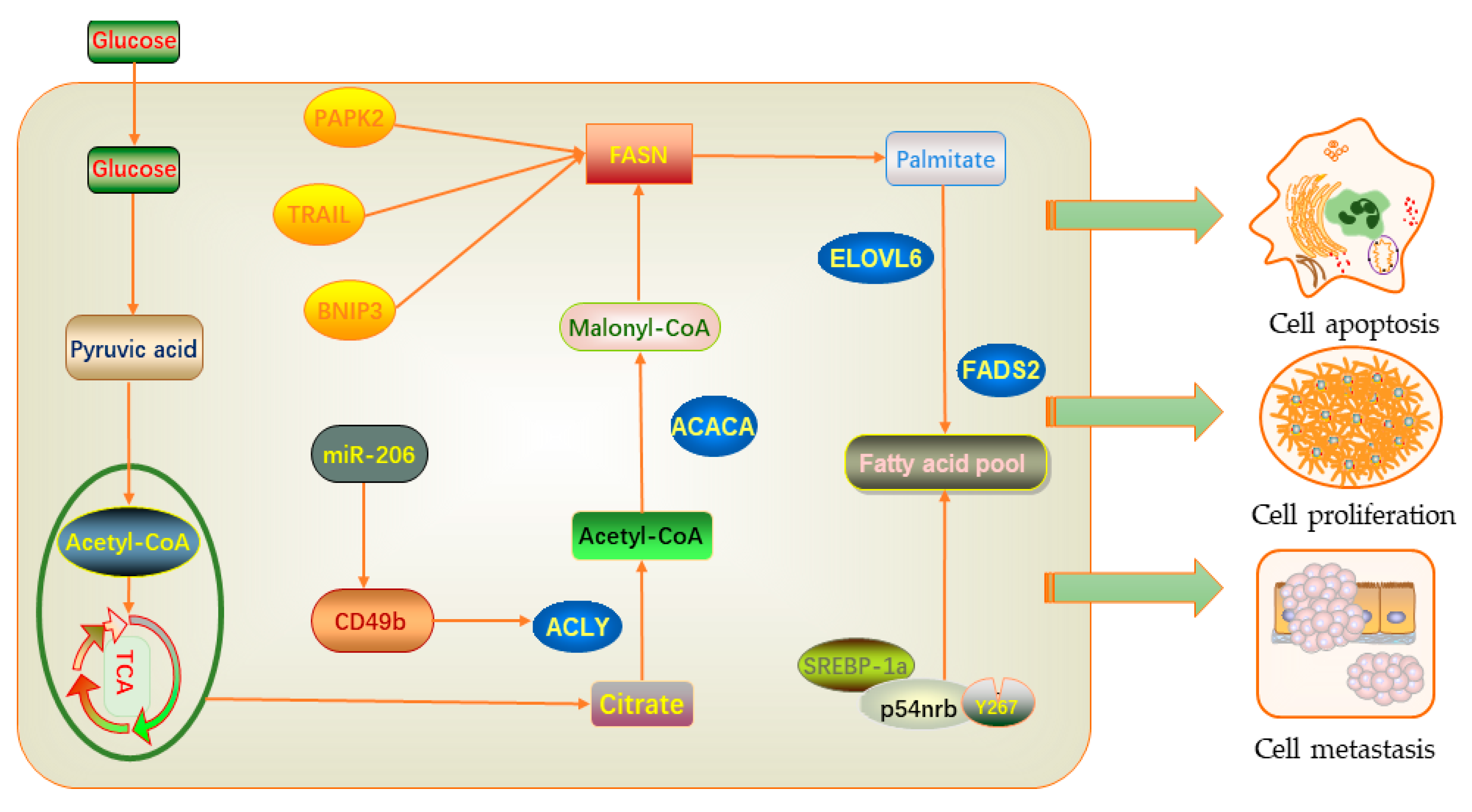
6. Effects of Lipid Metabolism on Proliferation, Migration, and Apoptosis in Breast Cancer
7. Lipids as Predictors of Breast Cancer Risk and Prognosis
8. LncRNA Related to Lipid Metabolism in Cancer
9. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- DeSantis, C.E.; Ma, J.; Gaudet, M.M.; Newman, L.A.; Miller, K.D.; Goding Sauer, A.; Jemal, A.; Siegel, R.L. Breast cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 438–451. [Google Scholar] [CrossRef]
- Paci, E.; Broeders, M.; Hofvind, S.; Puliti, D.; Duffy, S.W.; Group, E.W. European breast cancer service screening outcomes: A first balance sheet of the benefits and harms. Cancer Epidemiol. Biomark. Prev. 2014, 23, 1159–1163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McGuire, S. World Cancer Report 2014. Geneva, Switzerland: World Health Organization, International Agency for Research on Cancer, WHO Press, 2015. Adv. Nutr. 2016, 7, 418–419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pisano, E.D.; Gatsonis, C.; Hendrick, E.; Yaffe, M.; Baum, J.K.; Acharyya, S.; Conant, E.F.; Fajardo, L.L.; Bassett, L.; D’Orsi, C.; et al. Diagnostic performance of digital versus film mammography for breast-cancer screening. N. Engl. J. Med. 2005, 353, 1773–1783. [Google Scholar] [CrossRef] [PubMed]
- Zhi, H.; Xiao, X.Y.; Yang, H.Y.; Ou, B.; Wen, Y.L.; Luo, B.M. Ultrasonic elastography in breast cancer diagnosis: Strain ratio vs 5-point scale. Acad. Radiol. 2010, 17, 1227–1233. [Google Scholar] [CrossRef] [PubMed]
- Lehman, C.D.; Gatsonis, C.; Kuhl, C.K.; Hendrick, R.E.; Pisano, E.D.; Hanna, L.; Peacock, S.; Smazal, S.F.; Maki, D.D.; Julian, T.B.; et al. MRI evaluation of the contralateral breast in women with recently diagnosed breast cancer. N. Engl. J. Med. 2007, 356, 1295–1303. [Google Scholar] [CrossRef]
- Sechopoulos, I.; Teuwen, J.; Mann, R. Artificial intelligence for breast cancer detection in mammography and digital breast tomosynthesis: State of the art. Semin. Cancer Biol. 2020. [Google Scholar] [CrossRef]
- Birdwell, R.L.; Ikeda, D.M.; O’Shaughnessy, K.F.; Sickles, E.A. Mammographic characteristics of 115 missed cancers later detected with screening mammography and the potential utility of computer-aided detection. Radiology 2001, 219, 192–202. [Google Scholar] [CrossRef]
- Rotten, D.; Levaillant, J.M. The value of ultrasonic examination to detect and diagnose breast carcinomas. Analysis of the results obtained in 125 tumors using radiographic and ultrasound mammography. Ultrasound Obstet. Gynecol. 1992, 2, 203–214. [Google Scholar] [CrossRef]
- Olsen, O.; Gotzsche, P.C. Screening for breast cancer with mammography. Cochrane Database Syst. Rev. 2001. [Google Scholar] [CrossRef]
- Chen, X.; Chen, H.; Dai, M.; Ai, J.; Li, Y.; Mahon, B.; Dai, S.; Deng, Y. Plasma lipidomics profiling identified lipid biomarkers in distinguishing early-stage breast cancer from benign lesions. Oncotarget 2016, 7, 36622–36631. [Google Scholar] [CrossRef] [PubMed]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menendez, J.A.; Lupu, R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat. Rev. Cancer 2007, 7, 763–777. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ren, S.; Piao, H.L.; Wang, F.; Yin, P.; Xu, C.; Lu, X.; Ye, G.; Shao, Y.; Yan, M.; et al. Integration of lipidomics and transcriptomics unravels aberrant lipid metabolism and defines cholesteryl oleate as potential biomarker of prostate cancer. Sci. Rep. 2016, 6, 20984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merdad, A.; Karim, S.; Schulten, H.J.; Jayapal, M.; Dallol, A.; Buhmeida, A.; Al-Thubaity, F.; Gari, I.M.; Chaudhary, A.G.; Abuzenadah, A.M.; et al. Transcriptomics profiling study of breast cancer from Kingdom of Saudi Arabia revealed altered expression of Adiponectin and Fatty Acid Binding Protein4: Is lipid metabolism associated with breast cancer? BMC Genom. 2015, 16 (Suppl. S1), S11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christensen, L.L.; True, K.; Hamilton, M.P.; Nielsen, M.M.; Damas, N.D.; Damgaard, C.K.; Ongen, H.; Dermitzakis, E.; Bramsen, J.B.; Pedersen, J.S.; et al. SNHG16 is regulated by the Wnt pathway in colorectal cancer and affects genes involved in lipid metabolism. Mol. Oncol. 2016, 10, 1266–1282. [Google Scholar] [CrossRef] [PubMed]
- Jasbi, P.; Wang, D.; Cheng, S.L.; Fei, Q.; Cui, J.Y.; Liu, L.; Wei, Y.; Raftery, D.; Gu, H. Breast cancer detection using targeted plasma metabolomics. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2019, 1105, 26–37. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Li, H.Y.; Liao, H.W.; Lin, C.H.; Wang, C.Y.; Kuo, W.H.; Kuo, C.H. Using post-column infused internal standard assisted quantitative metabolomics for establishing prediction models for breast cancer detection. Rapid Commun. Mass Spectrom. 2020, 34 (Suppl. S1), e8581. [Google Scholar] [CrossRef]
- Yamashita, A.; Hayashi, Y.; Nemoto-Sasaki, Y.; Ito, M.; Oka, S.; Tanikawa, T.; Waku, K.; Sugiura, T. Acyltransferases and transacylases that determine the fatty acid composition of glycerolipids and the metabolism of bioactive lipid mediators in mammalian cells and model organisms. Prog. Lipid Res. 2014, 53, 18–81. [Google Scholar] [CrossRef]
- Li, L.; Zheng, X.; Zhou, Q.; Villanueva, N.; Nian, W.; Liu, X.; Huan, T. Metabolomics-Based Discovery of Molecular Signatures for Triple Negative Breast Cancer in Asian Female Population. Sci. Rep. 2020, 10, 370. [Google Scholar] [CrossRef]
- Li, X.; Liu, Z.L.; Wu, Y.T.; Wu, H.; Dai, W.; Arshad, B.; Xu, Z.; Li, H.; Wu, K.N.; Kong, L.Q. Status of lipid and lipoprotein in female breast cancer patients at initial diagnosis and during chemotherapy. Lipids Health Dis. 2018, 17, 91. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Bowers, J.; Liu, L.; Wei, S.; Gowda, G.A.; Hammoud, Z.; Raftery, D. Esophageal cancer metabolite biomarkers detected by LC-MS and NMR methods. PLoS ONE 2012, 7, e30181. [Google Scholar] [CrossRef]
- Gunther, U.L. Metabolomics Biomarkers for Breast Cancer. Pathobiology 2015, 82, 153–165. [Google Scholar] [CrossRef]
- Fahy, E.; Cotter, D.; Sud, M.; Subramaniam, S. Lipid classification, structures and tools. Biochim. Biophys. Acta 2011, 1811, 637–647. [Google Scholar] [CrossRef]
- Fahy, E.; Subramaniam, S.; Brown, H.A.; Glass, C.K.; Merrill, A.H., Jr.; Murphy, R.C.; Raetz, C.R.; Russell, D.W.; Seyama, Y.; Shaw, W.; et al. A comprehensive classification system for lipids. J. Lipid Res. 2005, 46, 839–861. [Google Scholar] [CrossRef] [Green Version]
- Fahy, E.; Subramaniam, S.; Murphy, R.C.; Nishijima, M.; Raetz, C.R.; Shimizu, T.; Spener, F.; van Meer, G.; Wakelam, M.J.; Dennis, E.A. Update of the LIPID MAPS comprehensive classification system for lipids. J. Lipid Res. 2009, 50, S9–S14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aro, A.; Mannisto, S.; Salminen, I.; Ovaskainen, M.L.; Kataja, V.; Uusitupa, M. Inverse association between dietary and serum conjugated linoleic acid and risk of breast cancer in postmenopausal women. Nutr. Cancer 2000, 38, 151–157. [Google Scholar] [CrossRef]
- Lands, W.E.; Hart, P. The control of fatty acid composition in glycerolipids. J. Am. Oil Chem. Soc. 1966, 43, 290–295. [Google Scholar] [CrossRef]
- Sankaram, M.B.; Thompson, T.E. Interaction of cholesterol with various glycerophospholipids and sphingomyelin. Biochemistry 1990, 29, 10670–10675. [Google Scholar] [CrossRef] [PubMed]
- Montealegre, C.; Verardo, V.; Luisa Marina, M.; Caboni, M.F. Analysis of glycerophospho- and sphingolipids by CE. Electrophoresis 2014, 35, 779–792. [Google Scholar] [CrossRef] [PubMed]
- Bartke, N.; Hannun, Y.A. Bioactive sphingolipids: Metabolism and function. J. Lipid Res. 2009, 50, S91–S96. [Google Scholar] [CrossRef] [Green Version]
- Chester, M.A. IUPAC-IUB Joint Commission on Biochemical Nomenclature (JCBN). Nomenclature of glycolipids--recommendations 1997. Eur. J. Biochem. 1998, 257, 293–298. [Google Scholar] [CrossRef] [PubMed]
- McDaniel, R.; Ebert-Khosla, S.; Hopwood, D.A.; Khosla, C. Engineered biosynthesis of novel polyketides. Science 1993, 262, 1546–1550. [Google Scholar] [CrossRef]
- Tsui-Pierchala, B.A.; Encinas, M.; Milbrandt, J.; Johnson, E.M. Lipid rafts in neuronal signaling and function. Trends Neurosci. 2002, 25, 412–417. [Google Scholar] [CrossRef]
- Wenk, M.R.; De Camilli, P. Protein-lipid interactions and phosphoinositide metabolism in membrane traffic: Insights from vesicle recycling in nerve terminals. Proc. Natl. Acad. Sci. USA 2004, 101, 8262–8269. [Google Scholar] [CrossRef] [Green Version]
- Heringdorf, D.M.Z.; Jakobs, K.H. Lysophospholipid receptors: Signalling, pharmacology and regulation by lysophospholipid metabolism. Biochim. Biophys. Acta 2007, 1768, 923–940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brinkmann, V. Sphingosine 1-phosphate receptors in health and disease: Mechanistic insights from gene deletion studies and reverse pharmacology. Pharmacol. Ther. 2007, 115, 84–105. [Google Scholar] [CrossRef]
- Beloribi-Djefaflia, S.; Vasseur, S.; Guillaumond, F. Lipid metabolic reprogramming in cancer cells. Oncogenesis 2016, 5, e189. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Gandhi, N.; Das, G. Metabolic Reprogramming in Breast Cancer and Its Therapeutic Implications. Cells 2019, 8, 89. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.L.; Sheng, R.; Jung, J.H.; Wang, L.; Stec, E.; O’Connor, M.J.; Song, S.; Bikkavilli, R.K.; Winn, R.A.; Lee, D.; et al. Orthogonal lipid sensors identify transbilayer asymmetry of plasma membrane cholesterol. Nat. Chem. Biol. 2017, 13, 268–274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hilvo, M.; Denkert, C.; Lehtinen, L.; Muller, B.; Brockmoller, S.; Seppanen-Laakso, T.; Budczies, J.; Bucher, E.; Yetukuri, L.; Castillo, S.; et al. Novel theranostic opportunities offered by characterization of altered membrane lipid metabolism in breast cancer progression. Cancer Res. 2011, 71, 3236–3245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, X.; Gross, R.W. Global analyses of cellular lipidomes directly from crude extracts of biological samples by ESI mass spectrometry: A bridge to lipidomics. J. Lipid Res. 2003, 44, 1071–1079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wenk, M.R. The emerging field of lipidomics. Nat. Rev. Drug Discov. 2005, 4, 594–610. [Google Scholar] [CrossRef] [PubMed]
- Myers, V.H.; Champagne, C.M. Nutritional effects on blood pressure. Curr. Opin. Lipidol. 2007, 18, 20–24. [Google Scholar] [CrossRef]
- Matyash, V.; Liebisch, G.; Kurzchalia, T.V.; Shevchenko, A.; Schwudke, D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J. Lipid Res. 2008, 49, 1137–1146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruiz-Gutiérrez, V.; Pérez-Camino, M.C. Update on solid-phase extraction for the analysis of lipid classes and related compounds. J. Chromatogr. A 2000, 885, 321–341. [Google Scholar] [CrossRef]
- Puranik, M.; Bhawsar, D.V.; Rathi, P.; Yeole, P.G. Simultaneous Determination of Ofloxacin and Ornidazole in Solid Dosage Form by RP-HPLC and HPTLC Techniques. Indian J. Pharm. Sci. 2010, 72, 513–517. [Google Scholar] [CrossRef] [Green Version]
- Taamalli, A.; Arraez-Roman, D.; Barrajon-Catalan, E.; Ruiz-Torres, V.; Perez-Sanchez, A.; Herrero, M.; Ibanez, E.; Micol, V.; Zarrouk, M.; Segura-Carretero, A.; et al. Use of advanced techniques for the extraction of phenolic compounds from Tunisian olive leaves: Phenolic composition and cytotoxicity against human breast cancer cells. Food Chem. Toxicol. 2012, 50, 1817–1825. [Google Scholar] [CrossRef] [Green Version]
- Rauser, S.; Marquardt, C.; Balluff, B.; Deininger, S.O.; Albers, C.; Belau, E.; Hartmer, R.; Suckau, D.; Specht, K.; Ebert, M.P.; et al. Classification of HER2 receptor status in breast cancer tissues by MALDI imaging mass spectrometry. J. Proteome Res. 2010, 9, 1854–1863. [Google Scholar] [CrossRef] [Green Version]
- Lubes, G.; Goodarzi, M. GC-MS based metabolomics used for the identification of cancer volatile organic compounds as biomarkers. J. Pharm. Biomed. Anal. 2018, 147, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Azab, S.; Ly, R.; Britz-McKibbin, P. Robust Method for High-Throughput Screening of Fatty Acids by Multisegment Injection-Nonaqueous Capillary Electrophoresis-Mass Spectrometry with Stringent Quality Control. Anal. Chem. 2019, 91, 2329–2336. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Zhang, Z.; Fu, X.; Li, W.; Wang, T.; Liu, H. Analysis of phospholipids by NACE with on-line ESI-MS. Electrophoresis 2007, 28, 1418–1425. [Google Scholar] [CrossRef] [PubMed]
- Cornett, D.S.; Mobley, J.A.; Dias, E.C.; Andersson, M.; Arteaga, C.L.; Sanders, M.E.; Caprioli, R.M. A novel histology-directed strategy for MALDI-MS tissue profiling that improves throughput and cellular specificity in human breast cancer. Mol. Cell Proteom. 2006, 5, 1975–1983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Z.; Bagarolo, G.I.; Thoroe-Boveleth, S.; Jankowski, J. “Lipidomics”: Mass spectrometric and chemometric analyses of lipids. Adv. Drug Deliv. Rev. 2020. [Google Scholar] [CrossRef] [PubMed]
- Bergers, G.; Hanahan, D. Modes of resistance to anti-angiogenic therapy. Nat. Rev. Cancer 2008, 8, 592–603. [Google Scholar] [CrossRef] [Green Version]
- Crawford, Y.; Kasman, I.; Yu, L.; Zhong, C.; Wu, X.; Modrusan, Z.; Kaminker, J.; Ferrara, N. PDGF-C mediates the angiogenic and tumorigenic properties of fibroblasts associated with tumors refractory to anti-VEGF treatment. Cancer Cell 2009, 15, 21–34. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.; Arbiser, J.; D’Amato, R.J.; D’Amore, P.A.; Ingber, D.E.; Kerbel, R.; Klagsbrun, M.; Lim, S.; Moses, M.A.; Zetter, B.; et al. Forty-year journey of angiogenesis translational research. Sci. Transl. Med. 2011, 3, 114rv113. [Google Scholar] [CrossRef]
- Iwamoto, H.; Abe, M.; Yang, Y.; Cui, D.; Seki, T.; Nakamura, M.; Hosaka, K.; Lim, S.; Wu, J.; He, X.; et al. Cancer Lipid Metabolism Confers Antiangiogenic Drug Resistance. Cell Metab. 2018, 28, 104–117.e105. [Google Scholar] [CrossRef] [Green Version]
- Jayashankar, V.; Edinger, A.L. Macropinocytosis confers resistance to therapies targeting cancer anabolism. Nat. Commun. 2020, 11, 1121. [Google Scholar] [CrossRef]
- Recouvreux, M.V.; Commisso, C. Macropinocytosis: A Metabolic Adaptation to Nutrient Stress in Cancer. Front. Endocrinol. (Lausanne) 2017, 8, 261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amyere, M.; Payrastre, B.; Krause, U.; Van Der Smissen, P.; Veithen, A.; Courtoy, P.J. Constitutive macropinocytosis in oncogene-transformed fibroblasts depends on sequential permanent activation of phosphoinositide 3-kinase and phospholipase C. Mol. Biol. Cell 2000, 11, 3453–3467. [Google Scholar] [CrossRef] [Green Version]
- Tautu, C.; Verazin, G.; Prorok, J.J.; Alhadeff, J.A. Improved procedure for determination of serum lipid-associated sialic acid: Application for early diagnosis of colorectal cancer. J. Natl. Cancer Inst. 1988, 80, 1333–1337. [Google Scholar] [CrossRef]
- Zeleznik, O.A.; Clish, C.B.; Kraft, P.; Avila-Pacheco, J.; Eliassen, A.H.; Tworoger, S.S. Circulating Lysophosphatidylcholines, Phosphatidylcholines, Ceramides, and Sphingomyelins and Ovarian Cancer Risk: A 23-Year Prospective Study. J. Natl. Cancer Inst. 2020, 112, 628–636. [Google Scholar] [CrossRef]
- Qiu, Y.; Zhou, B.; Su, M.; Baxter, S.; Zheng, X.; Zhao, X.; Yen, Y.; Jia, W. Mass spectrometry-based quantitative metabolomics revealed a distinct lipid profile in breast cancer patients. Int. J. Mol. Sci. 2013, 14, 8047–8061. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.; Min, H.K.; Kong, G.; Moon, M.H. Quantitative analysis of phosphatidylcholines and phosphatidylethanolamines in urine of patients with breast cancer by nanoflow liquid chromatography/tandem mass spectrometry. Anal. Bioanal. Chem. 2009, 393, 1649–1656. [Google Scholar] [CrossRef]
- Ide, Y.; Waki, M.; Hayasaka, T.; Nishio, T.; Morita, Y.; Tanaka, H.; Sasaki, T.; Koizumi, K.; Matsunuma, R.; Hosokawa, Y.; et al. Human breast cancer tissues contain abundant phosphatidylcholine(36ratio1) with high stearoyl-CoA desaturase-1 expression. PLoS ONE 2013, 8, e61204. [Google Scholar] [CrossRef]
- Iwano, T.; Yoshimura, K.; Inoue, S.; Odate, T.; Ogata, K.; Funatsu, S.; Tanihata, H.; Kondo, T.; Ichikawa, D.; Takeda, S. Breast cancer diagnosis based on lipid profiling by probe electrospray ionization mass spectrometry. Br. J. Surg. 2020, 107, 632–635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, N.; Zhang, G.; Pan, L.; Yan, C.; Zhang, L.; Weng, Y.; Wang, W.; Chen, X.; Yang, G. Potential plasma lipid biomarkers in early-stage breast cancer. Biotechnol. Lett. 2017, 39, 1657–1666. [Google Scholar] [CrossRef]
- Cifkova, E.; Holcapek, M.; Lisa, M.; Vrana, D.; Gatek, J.; Melichar, B. Determination of lipidomic differences between human breast cancer and surrounding normal tissues using HILIC-HPLC/ESI-MS and multivariate data analysis. Anal. Bioanal. Chem. 2015, 407, 991–1002. [Google Scholar] [CrossRef] [PubMed]
- Laisupasin, P.; Thompat, W.; Sukarayodhin, S.; Sornprom, A.; Sudjaroen, Y. Comparison of Serum Lipid Profiles between Normal Controls and Breast Cancer Patients. J. Lab. Physicians 2013, 5, 38–41. [Google Scholar] [CrossRef] [PubMed]
- Franky Dhaval, S.; Shilin Nandubhai, S.; Pankaj Manubhai, S.; Patel, H.R.; Prabhudas Shankerbhai, P. Significance of alterations in plasma lipid profile levels in breast cancer. Integr. Cancer Ther. 2008, 7, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Akalanka, H.M.K.; Ekanayake, S.; Samarasinghe, K. Could Anthropometric and Lipid Parameters Reflect Susceptibility to Breast Cancer? Comparison of Newly Diagnosed Breast Cancer and Apparently Healthy Women. Asian Pac. J. Cancer Prev. 2018, 19, 2475–2480. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Cui, X.; Zhang, N.; Li, M.; Bai, Y.; Han, X.; Shi, Y.; Liu, H. Comprehensive lipid profiling of plasma in patients with benign breast tumor and breast cancer reveals novel biomarkers. Anal. Bioanal. Chem. 2015, 407, 5065–5077. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Shin, Y.; Kim, T.H.; Kim, D.H.; Lee, A. Plasma metabolites as possible biomarkers for diagnosis of breast cancer. PLoS ONE 2019, 14, e0225129. [Google Scholar] [CrossRef]
- Brozek-Pluska, B.; Musial, J.; Kordek, R.; Bailo, E.; Dieing, T.; Abramczyk, H. Raman spectroscopy and imaging: Applications in human breast cancer diagnosis. Analyst 2012, 137, 3773–3780. [Google Scholar] [CrossRef]
- Zhang, Y.; Song, L.; Liu, N.; He, C.; Li, Z. Decreased serum levels of free fatty acids are associated with breast cancer. Clin. Chim. Acta 2014, 437, 31–37. [Google Scholar] [CrossRef]
- Lv, W.; Yang, T. Identification of possible biomarkers for breast cancer from free fatty acid profiles determined by GC-MS and multivariate statistical analysis. Clin. Biochem. 2012, 45, 127–133. [Google Scholar] [CrossRef]
- His, M.; Zelek, L.; Deschasaux, M.; Pouchieu, C.; Kesse-Guyot, E.; Hercberg, S.; Galan, P.; Latino-Martel, P.; Blacher, J.; Touvier, M. Prospective associations between serum biomarkers of lipid metabolism and overall, breast and prostate cancer risk. Eur. J. Epidemiol. 2014, 29, 119–132. [Google Scholar] [CrossRef]
- Yeo, W.; Mo, F.K.F.; Pang, E.; Suen, J.J.S.; Koh, J.; Loong, H.H.F.; Yip, C.C.H.; Ng, R.Y.W.; Yip, C.H.W.; Tang, N.L.S.; et al. Profiles of lipids, blood pressure and weight changes among premenopausal Chinese breast cancer patients after adjuvant chemotherapy. BMC Womens Health 2017, 17, 55. [Google Scholar] [CrossRef]
- Hammad, L.A.; Wu, G.; Saleh, M.M.; Klouckova, I.; Dobrolecki, L.E.; Hickey, R.J.; Schnaper, L.; Novotny, M.V.; Mechref, Y. Elevated levels of hydroxylated phosphocholine lipids in the blood serum of breast cancer patients. Rapid Commun. Mass Spectrom. 2009, 23, 863–876. [Google Scholar] [CrossRef]
- Min, H.K.; Kong, G.; Moon, M.H. Quantitative analysis of urinary phospholipids found in patients with breast cancer by nanoflow liquid chromatography-tandem mass spectrometry: II. Negative ion mode analysis of four phospholipid classes. Anal. Bioanal. Chem. 2010, 396, 1273–1280. [Google Scholar] [CrossRef]
- Chatgilialoglu, C.; Ferreri, C.; Melchiorre, M.; Sansone, A.; Torreggiani, A. Lipid geometrical isomerism: From chemistry to biology and diagnostics. Chem. Rev. 2014, 114, 255–284. [Google Scholar] [CrossRef]
- Kotronen, A.; Seppanen-Laakso, T.; Westerbacka, J.; Kiviluoto, T.; Arola, J.; Ruskeepaa, A.L.; Yki-Jarvinen, H.; Oresic, M. Comparison of lipid and fatty acid composition of the liver, subcutaneous and intra-abdominal adipose tissue, and serum. Obesity (Silver Spring) 2010, 18, 937–944. [Google Scholar] [CrossRef] [PubMed]
- Ferreri, C.; Sansone, A.; Ferreri, R.; Amezaga, J.; Tueros, I. Fatty Acids and Membrane Lipidomics in Oncology: A Cross-Road of Nutritional, Signaling and Metabolic Pathways. Metabolites 2020, 10, 345. [Google Scholar] [CrossRef] [PubMed]
- Kang, K.S.; Wang, P.; Yamabe, N.; Fukui, M.; Jay, T.; Zhu, B.T. Docosahexaenoic acid induces apoptosis in MCF-7 cells in vitro and in vivo via reactive oxygen species formation and caspase 8 activation. PLoS ONE 2010, 5, e10296. [Google Scholar] [CrossRef] [Green Version]
- Cao, W.; Ma, Z.; Rasenick, M.M.; Yeh, S.; Yu, J. N-3 poly-unsaturated fatty acids shift estrogen signaling to inhibit human breast cancer cell growth. PLoS ONE 2012, 7, e52838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiong, A.; Yu, W.; Tiwary, R.; Sanders, B.G.; Kline, K. Distinct roles of different forms of vitamin E in DHA-induced apoptosis in triple-negative breast cancer cells. Mol. Nutr. Food Res. 2012, 56, 923–934. [Google Scholar] [CrossRef] [PubMed]
- Sun-yong Hwang, T.-H.K. 2 Hae-Hyeog Lee,2 Heung Yeol Kim,3 and Juhyun Seo4. Effect of Docosahexaenoic Acid (DHA) on Breast Cancer Cells. Kosin Med. J. 2015, 30, 103. [Google Scholar] [CrossRef] [Green Version]
- Oresic, M.; Hanninen, V.A.; Vidal-Puig, A. Lipidomics: A new window to biomedical frontiers. Trends Biotechnol. 2008, 26, 647–652. [Google Scholar] [CrossRef]
- Hosokawa, Y.; Masaki, N.; Takei, S.; Horikawa, M.; Matsushita, S.; Sugiyama, E.; Ogura, H.; Shiiya, N.; Setou, M. Recurrent triple-negative breast cancer (TNBC) tissues contain a higher amount of phosphatidylcholine (32:1) than non-recurrent TNBC tissues. PLoS ONE 2017, 12, e0183724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bougnoux, P.; Hajjaji, N.; Maheo, K.; Couet, C.; Chevalier, S. Fatty acids and breast cancer: Sensitization to treatments and prevention of metastatic re-growth. Prog. Lipid Res. 2010, 49, 76–86. [Google Scholar] [CrossRef] [PubMed]
- Meikle, P.J.; Barlow, C.K.; Mellett, N.A.; Mundra, P.A.; Bonham, M.P.; Larsen, A.; Cameron-Smith, D.; Sinclair, A.; Nestel, P.J.; Wong, G. Postprandial Plasma Phospholipids in Men Are Influenced by the Source of Dietary Fat. J. Nutr. 2015, 145, 2012–2018. [Google Scholar] [CrossRef] [Green Version]
- Corbet, C.; Feron, O. Emerging roles of lipid metabolism in cancer progression. Curr. Opin. Clin. Nutr. Metab. Care 2017, 20, 254–260. [Google Scholar] [CrossRef] [PubMed]
- Corbet, C.; Pinto, A.; Martherus, R.; de Jesus, J.P.S.; Polet, F.; Feron, O. Acidosis Drives the Reprogramming of Fatty Acid Metabolism in Cancer Cells through Changes in Mitochondrial and Histone Acetylation. Cell Metab. 2016, 24, 311–323. [Google Scholar] [CrossRef] [Green Version]
- Furuta, E.; Pai, S.K.; Zhan, R.; Bandyopadhyay, S.; Watabe, M.; Mo, Y.Y.; Hirota, S.; Hosobe, S.; Tsukada, T.; Miura, K.; et al. Fatty acid synthase gene is up-regulated by hypoxia via activation of Akt and sterol regulatory element binding protein-1. Cancer Res. 2008, 68, 1003–1011. [Google Scholar] [CrossRef] [Green Version]
- Lu, S.; Archer, M.C. Sp1 coordinately regulates de novo lipogenesis and proliferation in cancer cells. Int. J. Cancer 2010, 126, 416–425. [Google Scholar] [CrossRef]
- Zeng, Y.; Ren, K.; Zhu, X.; Zheng, Z.; Yi, G. Long Noncoding RNAs: Advances in Lipid Metabolism. Adv. Clin. Chem. 2018, 87, 1–36. [Google Scholar] [CrossRef]
- Shav-Tal, Y.; Zipori, D. PSF and p54nrb/NonO-multi-functional nuclear proteins. FEBS Lett. 2002, 531, 109–114. [Google Scholar] [CrossRef] [Green Version]
- Pavao, M.; Huang, Y.H.; Hafer, L.J.; Moreland, R.B.; Traish, A.M. Immunodetection of nmt55/p54nrb isoforms in human breast cancer. BMC Cancer 2001, 1, 15. [Google Scholar] [CrossRef] [Green Version]
- Shao, W.; Espenshade, P.J. Expanding roles for SREBP in metabolism. Cell Metab. 2012, 16, 414–419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, X.; Zong, H.; Abdulla, A.; Yang, E.S.; Wang, Q.; Ji, J.Y.; Pessin, J.E.; Das, B.C.; Yang, F. Inhibition of SREBP transcriptional activity by a boron-containing compound improves lipid homeostasis in diet-induced obesity. Diabetes 2014, 63, 2464–2473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Z.; Zhao, X.; Zhao, L.; Yang, H.; Liu, L.; Li, J.; Wu, J.; Yang, F.; Huang, G.; Liu, J. p54(nrb)/NONO regulates lipid metabolism and breast cancer growth through SREBP-1A. Oncogene 2016, 35, 1399–1410. [Google Scholar] [CrossRef] [PubMed]
- Kuhajda, F.P. Fatty-acid synthase and human cancer: New perspectives on its role in tumor biology. Nutrition 2000, 16, 202–208. [Google Scholar] [CrossRef]
- Innocenzi, D.; Alo, P.L.; Balzani, A.; Sebastiani, V.; Silipo, V.; La Torre, G.; Ricciardi, G.; Bosman, C.; Calvieri, S. Fatty acid synthase expression in melanoma. J. Cutan. Pathol. 2003, 30, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Kusakabe, T.; Nashimoto, A.; Honma, K.; Suzuki, T. Fatty acid synthase is highly expressed in carcinoma, adenoma and in regenerative epithelium and intestinal metaplasia of the stomach. Histopathology 2002, 40, 71–79. [Google Scholar] [CrossRef]
- Adorno-Cruz, V.; Hoffmann, A.D.; Liu, X.; Dashzeveg, N.K.; Taftaf, R.; Wray, B.; Keri, R.A.; Liu, H. ITGA2 promotes expression of ACLY and CCND1 in enhancing breast cancer stemness and metastasis. Genes Dis. 2020. [Google Scholar] [CrossRef]
- Saikolappan, S.; Kumar, B.; Shishodia, G.; Koul, S.; Koul, H.K. Reactive oxygen species and cancer: A complex interaction. Cancer Lett. 2019, 452, 132–143. [Google Scholar] [CrossRef]
- Aggarwal, V.; Tuli, H.S.; Varol, A.; Thakral, F.; Yerer, M.B.; Sak, K.; Varol, M.; Jain, A.; Khan, M.A.; Sethi, G. Role of Reactive Oxygen Species in Cancer Progression: Molecular Mechanisms and Recent Advancements. Biomolecules 2019, 9, 735. [Google Scholar] [CrossRef] [Green Version]
- Liao, Z.; Chua, D.; Tan, N.S. Reactive oxygen species: A volatile driver of field cancerization and metastasis. Mol. Cancer 2019, 18, 65. [Google Scholar] [CrossRef] [PubMed]
- Rosa, D.; Catala, A. Fatty acid profiles and non enzymatic lipid peroxidation of microsomes and mitochondria from bovine liver, kidney, lung and heart. Arch. Physiol. Biochem. 1998, 106, 33–37. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, S.; Zhang, W.; Zhou, F.; Campbell, G.A.; Chan, L.L.; Thompson, E.B.; Ansari, N.H. Cellular lipid peroxidation end-products induce apoptosis in human lens epithelial cells. Free Radic. Biol. Med. 2002, 32, 360–369. [Google Scholar] [CrossRef]
- Agostinelli, E.; Seiler, N. Non-irradiation-derived reactive oxygen species (ROS) and cancer: Therapeutic implications. Amino Acids 2006, 31, 341–355. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.S. The signaling mechanism of ROS in tumor progression. Cancer Metastasis Rev. 2006, 25, 695–705. [Google Scholar] [CrossRef]
- Guicciardi, M.E.; Gores, G.J. Apoptosis: A mechanism of acute and chronic liver injury. Gut 2005, 54, 1024–1033. [Google Scholar] [CrossRef] [Green Version]
- Kitahara, C.M.; Berrington de Gonzalez, A.; Freedman, N.D.; Huxley, R.; Mok, Y.; Jee, S.H.; Samet, J.M. Total cholesterol and cancer risk in a large prospective study in Korea. J. Clin. Oncol. 2011, 29, 1592–1598. [Google Scholar] [CrossRef] [Green Version]
- Emaus, A.; Veierod, M.B.; Tretli, S.; Finstad, S.E.; Selmer, R.; Furberg, A.S.; Bernstein, L.; Schlichting, E.; Thune, I. Metabolic profile, physical activity, and mortality in breast cancer patients. Breast Cancer Res. Treat. 2010, 121, 651–660. [Google Scholar] [CrossRef]
- Eliassen, A.H.; Colditz, G.A.; Rosner, B.; Willett, W.C.; Hankinson, S.E. Serum lipids, lipid-lowering drugs, and the risk of breast cancer. Arch. Intern. Med. 2005, 165, 2264–2271. [Google Scholar] [CrossRef]
- Nelson, E.R.; Wardell, S.E.; Jasper, J.S.; Park, S.; Suchindran, S.; Howe, M.K.; Carver, N.J.; Pillai, R.V.; Sullivan, P.M.; Sondhi, V.; et al. 27-Hydroxycholesterol links hypercholesterolemia and breast cancer pathophysiology. Science 2013, 342, 1094–1098. [Google Scholar] [CrossRef] [Green Version]
- Kimbung, S.; Chang, C.Y.; Bendahl, P.O.; Dubois, L.; Thompson, J.W.; McDonnell, D.P.; Borgquist, S. Impact of 27-hydroxylase (CYP27A1) and 27-hydroxycholesterol in breast cancer. Endocr.-Relat. Cancer 2017, 24, 339–349. [Google Scholar] [CrossRef]
- Baek, A.E.; Yu, Y.A.; He, S.; Wardell, S.E.; Chang, C.Y.; Kwon, S.; Pillai, R.V.; McDowell, H.B.; Thompson, J.W.; Dubois, L.G.; et al. The cholesterol metabolite 27 hydroxycholesterol facilitates breast cancer metastasis through its actions on immune cells. Nat. Commun. 2017, 8, 864. [Google Scholar] [CrossRef] [PubMed]
- Baek, A.E.; Nelson, E.R. The Contribution of Cholesterol and Its Metabolites to the Pathophysiology of Breast Cancer. Horm. Cancer 2016, 7, 219–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chlebowski, R.T.; Anderson, G.L.; Gass, M.; Lane, D.S.; Aragaki, A.K.; Kuller, L.H.; Manson, J.E.; Stefanick, M.L.; Ockene, J.; Sarto, G.E.; et al. Estrogen plus progestin and breast cancer incidence and mortality in postmenopausal women. JAMA 2010, 304, 1684–1692. [Google Scholar] [CrossRef]
- Fuhrman, B.J.; Schairer, C.; Gail, M.H.; Boyd-Morin, J.; Xu, X.; Sue, L.Y.; Buys, S.S.; Isaacs, C.; Keefer, L.K.; Veenstra, T.D.; et al. Estrogen metabolism and risk of breast cancer in postmenopausal women. J. Natl. Cancer Inst. 2012, 104, 326–339. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Tang, H.; Wang, J.; Xie, X.; Liu, P.; Kong, Y.; Ye, F.; Shuang, Z.; Xie, Z.; Xie, X. The effect of preoperative serum triglycerides and high-density lipoprotein-cholesterol levels on the prognosis of breast cancer. Breast 2017, 32, 1–6. [Google Scholar] [CrossRef]
- Bahl, M.; Ennis, M.; Tannock, I.F.; Hux, J.E.; Pritchard, K.I.; Koo, J.; Goodwin, P.J. Serum lipids and outcome of early-stage breast cancer: Results of a prospective cohort study. Breast Cancer Res. Treat. 2005, 94, 135–144. [Google Scholar] [CrossRef]
- Ray, G.; Husain, S.A. Role of lipids, lipoproteins and vitamins in women with breast cancer. Clin. Biochem. 2001, 34, 71–76. [Google Scholar] [CrossRef]
- Ni, H.; Liu, H.; Gao, R. Serum Lipids and Breast Cancer Risk: A Meta-Analysis of Prospective Cohort Studies. PLoS ONE 2015, 10, e0142669. [Google Scholar] [CrossRef]
- Nowak, C.; Arnlov, J. A Mendelian randomization study of the effects of blood lipids on breast cancer risk. Nat. Commun. 2018, 9, 3957. [Google Scholar] [CrossRef] [Green Version]
- Kang, H.S.; Lee, S.C.; Park, Y.S.; Jeon, Y.E.; Lee, J.H.; Jung, S.Y.; Park, I.H.; Jang, S.H.; Park, H.M.; Yoo, C.W.; et al. Protein and lipid MALDI profiles classify breast cancers according to the intrinsic subtype. BMC Cancer 2011, 11, 465. [Google Scholar] [CrossRef] [Green Version]
- Mao, X.; He, J.; Li, T.; Lu, Z.; Sun, J.; Meng, Y.; Abliz, Z.; Chen, J. Application of imaging mass spectrometry for the molecular diagnosis of human breast tumors. Sci. Rep. 2016, 6, 21043. [Google Scholar] [CrossRef] [PubMed]
- Escande, C.; Chini, C.C.; Nin, V.; Dykhouse, K.M.; Novak, C.M.; Levine, J.; van Deursen, J.; Gores, G.J.; Chen, J.; Lou, Z.; et al. Deleted in breast cancer-1 regulates SIRT1 activity and contributes to high-fat diet-induced liver steatosis in mice. J. Clin. Investig. 2010, 120, 545–558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hilakivi-Clarke, L.; Clarke, R.; Onojafe, I.; Raygada, M.; Cho, E.; Lippman, M. A maternal diet high in n-6 polyunsaturated fats alters mammary gland development, puberty onset, and breast cancer risk among female rat offspring. Proc. Natl. Acad. Sci. USA 1997, 94, 9372–9377. [Google Scholar] [CrossRef] [Green Version]
- Lupien, L.E.; Bloch, K.; Dehairs, J.; Traphagen, N.A.; Feng, W.W.; Davis, W.L.; Dennis, T.; Swinnen, J.V.; Wells, W.A.; Smits, N.C.; et al. Endocytosis of very low-density lipoproteins: An unexpected mechanism for lipid acquisition by breast cancer cells. J. Lipid Res. 2020, 61, 205–218. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Yao, J.; Chen, Y.; Geng, P.; Zhang, H.; Ma, X.; Zhao, J.; Yu, X. Expression and Functional Role of Reprogramming-Related Long Noncoding RNA (lincRNA-ROR) in Glioma. J. Mol. Neurosci. 2015, 56, 623–630. [Google Scholar] [CrossRef]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef]
- Panzitt, K.; Tschernatsch, M.M.; Guelly, C.; Moustafa, T.; Stradner, M.; Strohmaier, H.M.; Buck, C.R.; Denk, H.; Schroeder, R.; Trauner, M.; et al. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology 2007, 132, 330–342. [Google Scholar] [CrossRef]
- Seyed Hosseyni, M.; Gholamin, A.; Roohollah, F.; Sadeghizadeh, M. Evaluation of Expression Levels of Linc-ROR and HULC Genes in Breast Cancer Cells (MCF7) Following Treatment with Nanocurcumin. J. Hum. Genet. Genom. 2020, 3. [Google Scholar] [CrossRef]
- Hildebrandt, M.A.; Gu, J.; Lin, J.; Ye, Y.; Tan, W.; Tamboli, P.; Wood, C.G.; Wu, X. Hsa-miR-9 methylation status is associated with cancer development and metastatic recurrence in patients with clear cell renal cell carcinoma. Oncogene 2010, 29, 5724–5728. [Google Scholar] [CrossRef] [Green Version]
- Phillips, C.M.; Goumidi, L.; Bertrais, S.; Field, M.R.; Cupples, L.A.; Ordovas, J.M.; Defoort, C.; Lovegrove, J.A.; Drevon, C.A.; Gibney, M.J.; et al. Gene-nutrient interactions with dietary fat modulate the association between genetic variation of the ACSL1 gene and metabolic syndrome. J. Lipid Res. 2010, 51, 1793–1800. [Google Scholar] [CrossRef] [Green Version]
- Ong, K.T.; Mashek, M.T.; Bu, S.Y.; Greenberg, A.S.; Mashek, D.G. Adipose triglyceride lipase is a major hepatic lipase that regulates triacylglycerol turnover and fatty acid signaling and partitioning. Hepatology 2011, 53, 116–126. [Google Scholar] [CrossRef] [Green Version]
- Mashek, D.G.; Bornfeldt, K.E.; Coleman, R.A.; Berger, J.; Bernlohr, D.A.; Black, P.; DiRusso, C.C.; Farber, S.A.; Guo, W.; Hashimoto, N.; et al. Revised nomenclature for the mammalian long-chain acyl-CoA synthetase gene family. J. Lipid Res. 2004, 45, 1958–1961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Cai, X.; Zhang, S.; Cui, M.; Liu, F.; Sun, B.; Zhang, W.; Zhang, X.; Ye, L. HBXIP up-regulates ACSL1 through activating transcriptional factor Sp1 in breast cancer. Biochem. Biophys. Res. Commun. 2017, 484, 565–571. [Google Scholar] [CrossRef]
- Lu, W.; Cao, F.; Wang, S.; Sheng, X.; Ma, J. LncRNAs: The Regulator of Glucose and Lipid Metabolism in Tumor Cells. Front. Oncol. 2019, 9, 1099. [Google Scholar] [CrossRef]
- Lan, X.; Yan, J.; Ren, J.; Zhong, B.; Li, J.; Li, Y.; Liu, L.; Yi, J.; Sun, Q.; Yang, X.; et al. A novel long noncoding RNA Lnc-HC binds hnRNPA2B1 to regulate expressions of Cyp7a1 and Abca1 in hepatocytic cholesterol metabolism. Hepatology 2016, 64, 58–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, J.; Feng, J.; Zhou, X. Long non-coding RNA HAGLROS regulates lipid metabolism reprogramming in intrahepatic cholangiocarcinoma via the mTOR signaling pathway. Exp. Mol. Pathol. 2020, 115, 104466. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Liang, B.; Wang, C.; Li, S.; Zhao, Y.; Huang, Q.; Liu, Z.; Yao, Z.; Wu, Q.; Liao, W.; et al. MSC-regulated lncRNA MACC1-AS1 promotes stemness and chemoresistance through fatty acid oxidation in gastric cancer. Oncogene 2019, 38, 4637–4654. [Google Scholar] [CrossRef] [Green Version]
- Cui, M.; Xiao, Z.; Wang, Y.; Zheng, M.; Song, T.; Cai, X.; Sun, B.; Ye, L.; Zhang, X. Long noncoding RNA HULC modulates abnormal lipid metabolism in hepatoma cells through an miR-9-mediated RXRA signaling pathway. Cancer Res. 2015, 75, 846–857. [Google Scholar] [CrossRef] [Green Version]
- Shang, C.; Wang, W.; Liao, Y.; Chen, Y.; Liu, T.; Du, Q.; Huang, J.; Liang, Y.; Liu, J.; Zhao, Y.; et al. LNMICC Promotes Nodal Metastasis of Cervical Cancer by Reprogramming Fatty Acid Metabolism. Cancer Res. 2018, 78, 877–890. [Google Scholar] [CrossRef] [Green Version]
- Sallam, T.; Jones, M.C.; Gilliland, T.; Zhang, L.; Wu, X.; Eskin, A.; Sandhu, J.; Casero, D.; Vallim, T.Q.; Hong, C.; et al. Feedback modulation of cholesterol metabolism by the lipid-responsive non-coding RNA LeXis. Nature 2016, 534, 124–128. [Google Scholar] [CrossRef] [Green Version]
- Li, D.; Cheng, M.; Niu, Y.; Chi, X.; Liu, X.; Fan, J.; Fan, H.; Chang, Y.; Yang, W. Identification of a novel human long non-coding RNA that regulates hepatic lipid metabolism by inhibiting SREBP-1c. Int. J. Biol. Sci. 2017, 13, 349–357. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Liu, B.; Chen, Z.; Li, G.; Zhang, Z. MSC-induced lncRNA HCP5 drove fatty acid oxidation through miR-3619-5p/AMPK/PGC1alpha/CEBPB axis to promote stemness and chemo-resistance of gastric cancer. Cell Death Dis. 2020, 11, 233. [Google Scholar] [CrossRef] [Green Version]
- Yan, C.; Chen, J.; Chen, N. Long noncoding RNA MALAT1 promotes hepatic steatosis and insulin resistance by increasing nuclear SREBP-1c protein stability. Sci. Rep. 2016, 6, 22640. [Google Scholar] [CrossRef] [PubMed]

| Diagnosis Biomarker | Sample Type | Lipidomics Technique | Reference |
|---|---|---|---|
| PCs, ePC (38:3) and ePC (32:2), PC ae C42:5 and PC aa C42:2 | Urine, Plasma, Tissue | nLC-ESI-MS-MS, ESI-MS/MS, UPLC-MS | [11,42,65,66] |
| PC (32:1), (34:4), (38:3), (40:5), (40:3), (44:11) | Plasma, Tissue | ESI-MS/MS, MALDI-IMS | [11,67] |
| PC (30:0), (34:0), (34:1), (36:0), (36:1), (36:2), (38:4), (40:6), (42:6) | Tissue | MALDI-IMS, probe electrospray ionization-MS | [67,68] |
| PC (20:2/20:5) and PC (20:0/24:1;) | Plasma | UPLC-quadrupole time-of-flight tandem/MS | [69] |
| LPC (18:0), (18:3), (20:0), (20:1), (20:2) and a C (16:0) | Plasma, Urine, Tissue | ESI-MS/MS, MALDI-IMS, nLC-ESI-MS-MS | [11,65,67] |
| PEs and PE (15:0/19:1) | Plasma, Tissue, Urine | UPLC-quadrupole time-of-flight tandem/MS, HILIC-HPLC/ ESI-MS, nLC-ESI-MS-MS | [66,69,70] |
| Triglyceride, Triglyceride (12:0/14:1) | Plasma | UPLC-quadrupole time-of-flight tandem/MS | [69,71,72,73] |
| Diglyceride (18:1/18:2) | Plasma | UPLC-quadrupole time-of-flight tandem/MS | [69] |
| C19:0 CE, C19:1 CE, C19:2 CE | Plasma | ESI-MS/MS | [11] |
| PI (16L:0/16:1) and PI (18:0/20:4) | Plasma | normal-phase/reversed-phase two-dimensional LC-MS | [74] |
| Docosahexaenoic acid | Plasma, Tissue | LC-MS, Raman spectroscopy | [75,76] |
| Fatty acids: C14:0, C16:0, C16:1, C18:0, C18:3, C18:2, C20:4, and C22:6 | Serum | Chip-based direct-infusion nano-electrospray ionization-Fourier transform ion cyclotron resonance mass spectrometry, GC–MS | [76,77,78] |
| HDL-C, VLDL-C, LDL-C, TC | Plasma | NA | [72,79,80] |
| N-palmitoyl proline | Plasma | UPLC-quadrupole time-of-flight tandem/MS | [69] |
| LncRNAs | Lipid Metabolism-Related Genes | Cancer Types | Influences | References |
|---|---|---|---|---|
| HC | hnRNPA2B1 | Hepatic carcinoma | TG and CHO decreased | [145] |
| HAGLROS | FASN, ACC, SCD1, CPT1, SREBP1, PPARγ | Intrahepatic cholangiocarcinoma | Promote autophagy, improving lipid metabolism reprogramming | [146] |
| MACC1-AS1 | ACS, CPT1A | Gastric cancer | Drug resistance | [147] |
| HULC | ACSL1, PPARA | Hepatic carcinoma | TG and CHO accumulated | [148] |
| LNMICC | FABP5 | Cervical cancer | Lymph node metastasis | [149] |
| LeXis | RALY | Hepatic carcinoma | CHO increased | [150] |
| HR1 | SREBP-1c, FASN | Hepatic carcinoma | TG and lipid droplet accumulated | [151] |
| HCP5 | CPT1 | Gastric cancer | Drug resistance | [152] |
| MALAT1 | SREBP-1c | Hepatic carcinoma | Hepatic steatosis and insulin resistance | [153] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, R.; Chen, Y.; Borgard, H.; Jijiwa, M.; Nasu, M.; He, M.; Deng, Y. The Function and Mechanism of Lipid Molecules and Their Roles in The Diagnosis and Prognosis of Breast Cancer. Molecules 2020, 25, 4864. https://doi.org/10.3390/molecules25204864
Guo R, Chen Y, Borgard H, Jijiwa M, Nasu M, He M, Deng Y. The Function and Mechanism of Lipid Molecules and Their Roles in The Diagnosis and Prognosis of Breast Cancer. Molecules. 2020; 25(20):4864. https://doi.org/10.3390/molecules25204864
Chicago/Turabian StyleGuo, Rui, Yu Chen, Heather Borgard, Mayumi Jijiwa, Masaki Nasu, Min He, and Youping Deng. 2020. "The Function and Mechanism of Lipid Molecules and Their Roles in The Diagnosis and Prognosis of Breast Cancer" Molecules 25, no. 20: 4864. https://doi.org/10.3390/molecules25204864
APA StyleGuo, R., Chen, Y., Borgard, H., Jijiwa, M., Nasu, M., He, M., & Deng, Y. (2020). The Function and Mechanism of Lipid Molecules and Their Roles in The Diagnosis and Prognosis of Breast Cancer. Molecules, 25(20), 4864. https://doi.org/10.3390/molecules25204864







