In Silico Molecular Study of Tryptophan Bitterness
Abstract
:1. Introduction
2. Results
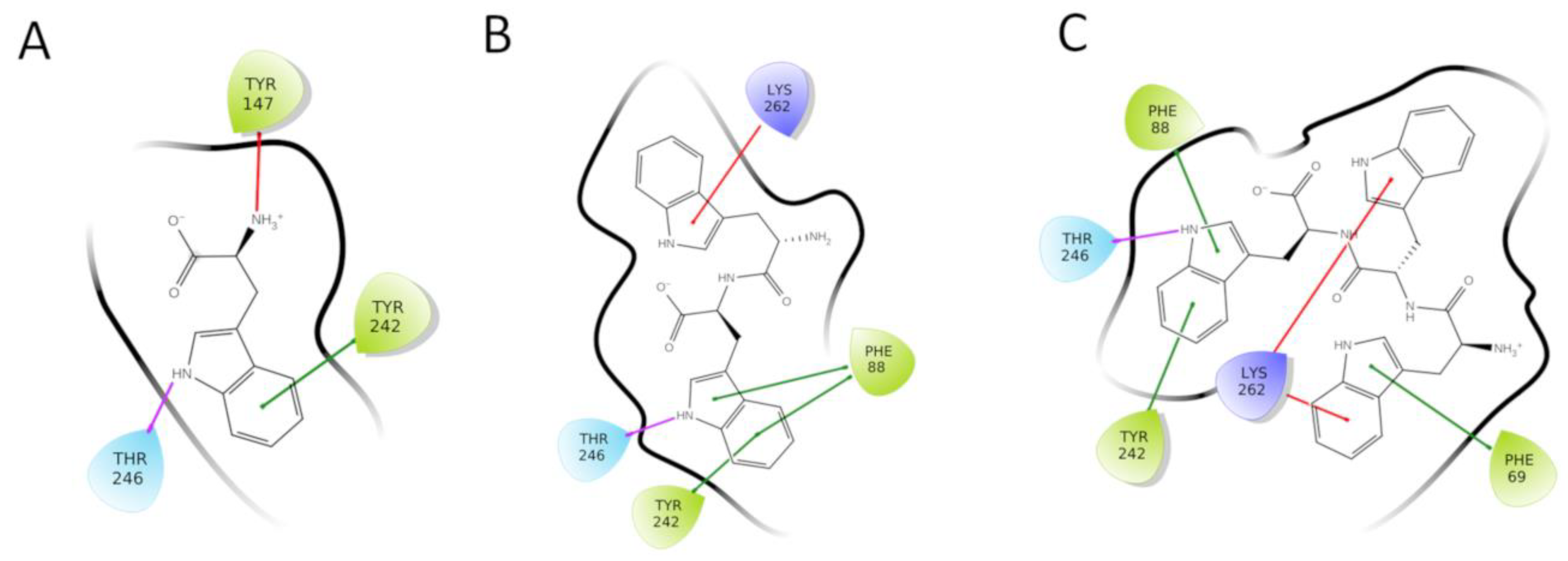
2.1. From the Promiscuous Tri-Tryptophan to the Selective Tryptophan
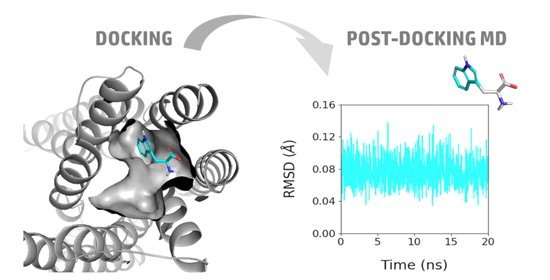
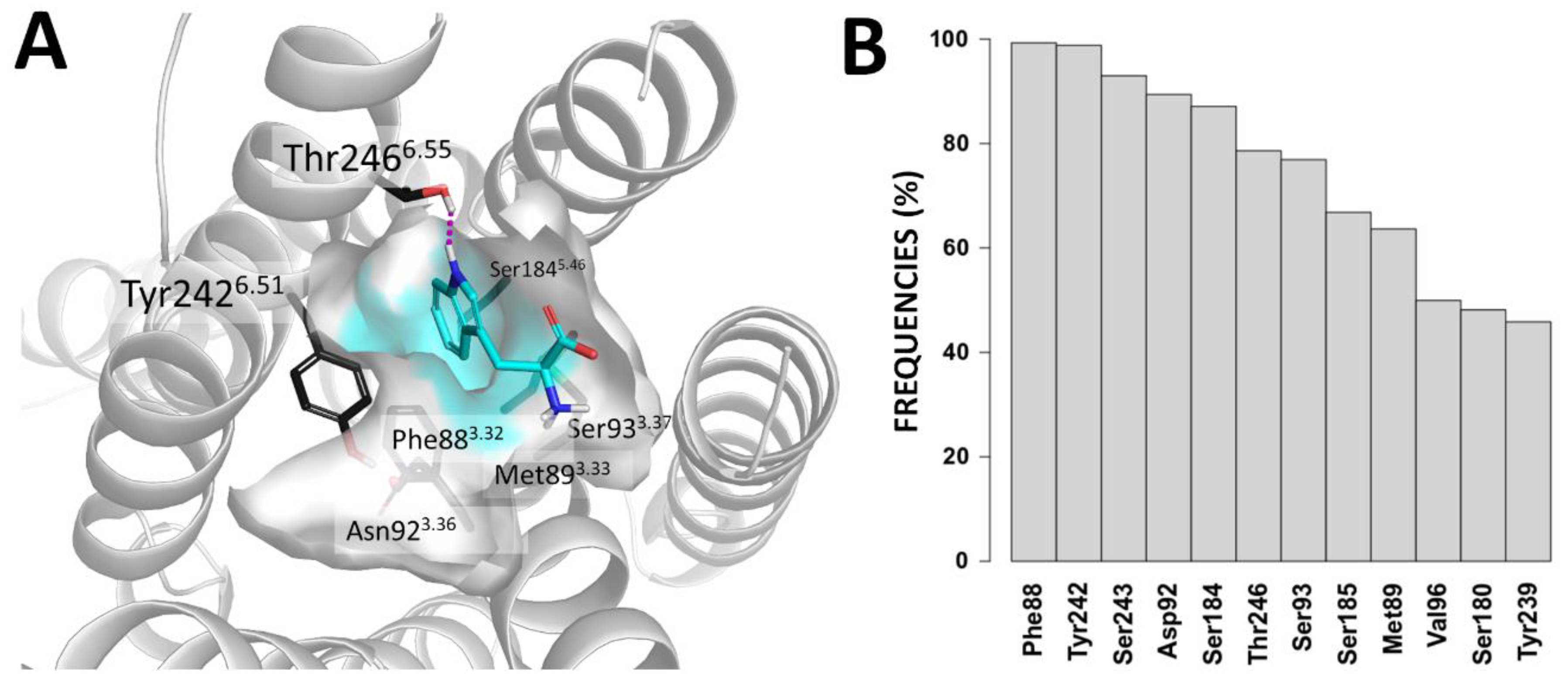
2.2. Predicted Binding Mode of Tryptophan into TAS2R4
3. Discussion
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jenkins, T.A.; Nguyen, J.C.; Polglaze, K.E.; Bertrand, P.P. Influence of tryptophan and serotonin on mood and cognition with a possible role of the gut-brain axis. Nutrients 2016, 8, 56. [Google Scholar] [CrossRef] [PubMed]
- Richard, D.M.; Dawes, M.A.; Mathias, C.W.; Acheson, A.; Hill-Kapturczak, N.; Dougherty, D.M. l-Tryptophan: Basic metabolic functions, behavioral research and therapeutic indications. Int. J. Tryptophan Res. 2009, 2, 45–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaluzna-Czaplinska, J.; Gatarek, P.; Chirumbolo, S.; Chartrand, M.S.; Bjorklund, G. How important is tryptophan in human health? Crit. Rev. Food Sci. 2019, 59, 72–88. [Google Scholar] [CrossRef] [PubMed]
- Nikolaus, S.; Schulte, B.; Al-Massad, N.; Thieme, F.; Schulte, D.M.; Bethge, J.; Rehman, A.; Tran, F.; Aden, K.; Hasler, R.; et al. Increased tryptophan metabolism is associated with activity of inflammatory bowel diseases. Gastroenterology 2017, 153, 1504–1516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Comai, S.; Bertazzo, A.; Brughera, M.; Crotti, S. Tryptophan in health and disease. Adv. Clin. Chem. 2020, 95, 165–218. [Google Scholar] [CrossRef] [PubMed]
- Wieser, H.; Belitz, H.D. Relations between structure and bitter taste of amino-acids and peptides. 1. Amino-acids and related compounds. Z. Lebensm.-Unters. Forsch. 1975, 159, 65–72. [Google Scholar] [CrossRef]
- Drewnowski, A.; Gomez-Carneros, C. Bitter taste, phytonutrients, and the consumer: A review. Am. J. Clin. Nutr. 2000, 72, 1424–1435. [Google Scholar] [CrossRef]
- Delompre, T.; Guichard, E.; Briand, L.; Salles, C. Taste perception of nutrients found in nutritional supplements: A review. Nutrients 2019, 11, 2050. [Google Scholar] [CrossRef] [Green Version]
- Adler, E.; Hoon, M.A.; Mueller, K.L.; Chandrashekar, J.; Ryba, N.J.; Zuker, C.S. A novel family of mammalian taste receptors. Cell 2000, 100, 693–702. [Google Scholar] [CrossRef] [Green Version]
- Chandrashekar, J.; Mueller, K.L.; Hoon, M.A.; Adler, E.; Feng, L.X.; Guo, W.; Zuker, C.S.; Ryba, N.J.P. T2Rs function as bitter taste receptors. Cell 2000, 100, 703–711. [Google Scholar] [CrossRef] [Green Version]
- Matsunami, H.; Montmayeur, J.P.; Buck, L.B. A family of candidate taste receptors in human and mouse. Nature 2000, 404, 601–604. [Google Scholar] [CrossRef] [PubMed]
- Di Pizio, A.; Niv, M.Y. Computational Studies of Smell and Taste Receptors. Isr. J. Chem. 2014, 54, 1205–1218. [Google Scholar] [CrossRef]
- Di Pizio, A.; Levit, A.; Slutzki, M.; Behrens, M.; Karaman, R.; Niv, M.Y. Comparing Class A GPCRs to bitter taste receptors: Structural motifs, ligand interactions and agonist-to-antagonist ratios. Method Cell Biol. 2016, 132, 401–427. [Google Scholar] [CrossRef]
- Di Pizio, A.; Niv, M.Y. Promiscuity and selectivity of bitter molecules and their receptors. Bioorgan. Med. Chem. 2015, 23, 4082–4091. [Google Scholar] [CrossRef]
- Toelstede, S.; Hofmann, T. Quantitative studies and taste re-engineering experiments toward the decoding of the nonvolatile sensometabolome of Gouda cheese. J. Agr. Food Chem. 2008, 56, 5299–5307. [Google Scholar] [CrossRef]
- Kohl, S.; Behrens, M.; Dunkel, A.; Hofmann, T.; Meyerhof, W. Amino acids and peptides activate at least five members of the human bitter taste receptor family. J. Agr. Food Chem. 2013, 61, 53–60. [Google Scholar] [CrossRef]
- Lee, S.J.; Depoortere, I.; Hatt, H. Therapeutic potential of ectopic olfactory and taste receptors. Nat. Rev. Drug Discov. 2019, 18, 116–138. [Google Scholar] [CrossRef]
- Di Pizio, A.; Behrens, M.; Krautwurst, D. Beyond the Flavour: The Potential Druggability of Chemosensory G Protein-Coupled Receptors. Int. J. Mol. Sci. 2019, 20, 1402. [Google Scholar] [CrossRef] [Green Version]
- Spaggiari, G.; Di Pizio, A.; Cozzini, P. Sweet, umami and bitter taste receptors: State of the art of in silico molecular modeling approaches. Trends Food Sci. Tech. 2020, 96, 21–29. [Google Scholar] [CrossRef]
- Upadhyaya, J.; Pydi, S.P.; Singh, N.; Aluko, R.E.; Chelikani, P. Bitter taste receptor T2R1 is activated by dipeptides and tripeptides. Biochem. Bioph. Res. Commun. 2010, 398, 331–335. [Google Scholar] [CrossRef]
- Dai, W.M.; You, Z.L.; Zhou, H.; Zhang, J.; Hu, Y.Q. Structure-function relationships of the human bitter taste receptor hTAS2R1: Insights from molecular modeling studies. J. Recept. Signal Transduct. 2011, 31, 229–240. [Google Scholar] [CrossRef] [PubMed]
- Stoeger, V.; Holik, A.K.; Holz, K.; Dingjan, T.; Hans, J.; Ley, J.P.; Krammer, G.E.; Niv, M.Y.; Somoza, M.M.; Somoza, V. Bitter-tasting amino acids l-arginine and l-isoleucine differentially regulate proton secretion via t2r1 signaling in human parietal cells in culture. J. Agr. Food Chem. 2020, 68, 3434–3444. [Google Scholar] [CrossRef] [PubMed]
- Born, S.; Levit, A.; Niv, M.Y.; Meyerhof, W.; Behrens, M. The human bitter taste receptor TAS2R10 is tailored to accommodate numerous diverse ligands. J. Neurosci. 2013, 33, 201–213. [Google Scholar] [CrossRef] [PubMed]
- Brockhoff, A.; Behrens, M.; Niv, M.Y.; Meyerhof, W. Structural requirements of bitter taste receptor activation. Proc. Natl. Acad. Sci. USA 2010, 107, 11110–11115. [Google Scholar] [CrossRef] [Green Version]
- Nowak, S.; Di Pizio, A.; Levit, A.; Niv, M.Y.; Meyerhof, W.; Behrens, M. Reengineering the ligand sensitivity of the broadly tuned human bitter taste receptor TAS2R14. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 2162–2173. [Google Scholar] [CrossRef]
- Shoichet, B.K.; Kobilka, B.K. Structure-based drug screening for G-protein-coupled receptors. Trends Pharmacol. Sci. 2012, 33, 268–272. [Google Scholar] [CrossRef] [Green Version]
- Di Pizio, A.; Kruetzfeldt, L.M.; Cheled-Shoval, S.; Meyerhof, W.; Behrens, M.; Niv, M.Y. Ligand binding modes from low resolution GPCR models and mutagenesis: Chicken bitter taste receptor as a test-case. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef]
- Di Pizio, A.; Waterloo, L.A.W.; Brox, R.; Lober, S.; Weikert, D.; Behrens, M.; Gmeiner, P.; Niv, M.Y. Rational design of agonists for bitter taste receptor TAS2R14: From modeling to bench and back. Cell Mol. Life Sci. 2019, 77, 531–542. [Google Scholar] [CrossRef]
- Ballesteros, J.A.; Weinstein, H. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods Neurosci. 1995, 25, 366–428. [Google Scholar]
- Dunkel, A.; Hofmann, T.; Di Pizio, A. In silico investigation of bitter hop-derived compounds and their cognate bitter taste receptors. J. Agric. Food Chem. 2020, 68, 10414–10423. [Google Scholar] [CrossRef]
- Sandal, M.; Behrens, M.; Brockhoff, A.; Musiani, F.; Giorgetti, A.; Carloni, P.; Meyerhof, W. Evidence for a transient additional ligand binding site in the tas2r46 bitter taste receptor. J. Chem. Theory Comput. 2015, 11, 4439–4449. [Google Scholar] [CrossRef] [PubMed]
- Fierro, F.; Giorgetti, A.; Carloni, P.; Meyerhof, W.; Alfonso-Prieto, M. Dual binding mode of “bitter sugars” to their human bitter taste receptor target. Sci. Rep. 2019, 9, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marchiori, A.; Capece, L.; Giorgetti, A.; Gasparini, P.; Behrens, M.; Carloni, P.; Meyerhof, W. Coarse-grained/molecular mechanics of the tas2r38 bitter taste receptor: Experimentally-validated detailed structural prediction of agonist binding. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakurai, T.; Misaka, T.; Ishiguro, M.; Masuda, K.; Sugawara, T.; Ito, K.; Kobayashi, T.; Matsuo, S.; Ishimaru, Y.; Asakura, T.; et al. Characterization of the β-d-Glucopyranoside binding site of the human bitter taste receptor hTAS2R16. J. Biol. Chem. 2010, 285, 28373–28378. [Google Scholar] [CrossRef] [Green Version]
- Xue, A.Y.; Di Pizio, A.; Levit, A.; Yarnitzky, T.; Penn, O.; Pupko, T.; Niv, M.Y. Independent evolution of strychnine recognition by bitter taste receptor subtypes. Front. Mol. Biosci. 2018, 5, 9. [Google Scholar] [CrossRef] [Green Version]
- Sherman, W.; Day, T.; Jacobson, M.P.; Friesner, R.A.; Farid, R. Novel procedure for modeling ligand/receptor induced fit effects. J. Med. Chem. 2006, 49, 534–553. [Google Scholar] [CrossRef]
- Pydi, S.P.; Sobotkiewicz, T.; Billakanti, R.; Bhullar, R.P.; Loewen, M.C.; Chelikani, P. Amino acid derivatives as bitter taste receptor (T2R) Blockers. J. Biol. Chem. 2014, 289, 25054–25066. [Google Scholar] [CrossRef] [Green Version]
- Meyerhof, W.; Batram, C.; Kuhn, C.; Brockhoff, A.; Chudoba, E.; Bufe, B.; Appendino, G.; Behrens, M. The molecular receptive ranges of human tas2r bitter taste receptors. Chem. Senses 2010, 35, 157–170. [Google Scholar] [CrossRef]
- Roland, W.S.U.; Vincken, J.P.; Gouka, R.J.; van Buren, L.; Gruppen, H.; Smit, G. Soy isoflavones and other isoflavonoids activate the human bitter taste receptors hTAS2R14 and hTAS2R39. J. Agr. Food Chem. 2011, 59, 11764–11771. [Google Scholar] [CrossRef]
- Tubert-Brohman, I.; Sherman, W.; Repasky, M.; Beuming, T. Improved docking of polypeptides with Glide. J. Chem. Inf. Modeling 2013, 53, 1689–1699. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD—Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Lomize, M.A.; Lomize, A.L.; Pogozheva, I.D.; Mosberg, H.I. OPM: Orientations of proteins in membranes database. Bioinformatics 2006, 22, 623–625. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926. [Google Scholar] [CrossRef]
- Harvey, M.J.; Giupponi, G.; Fabritiis, G.D. ACEMD: Accelerating biomolecular dynamics in the microsecond time scale. J. Chem. Theory Comput. 2009, 5, 1632–1639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, J.; MacKerell, A.D. CHARMM36 All-atom additive protein force field: Validation based on comparison to NMR data. J. Comput. Chem. 2013, 34, 2135–2145. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; de Groot, B.L.; Grubmüller, H.; MacKerell, A.D. CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods 2017, 14, 71–73. [Google Scholar] [CrossRef] [Green Version]
- Forester, T.R.; Smith, W. SHAKE, rattle and roll: Efficient constraint algorithms for linked rigid bodies. J. Comput. Chem. 1998, 19, 102–111. [Google Scholar] [CrossRef] [Green Version]
- Kräutler, V.; van Gunsteren, W.F.; Hünenberger, P.H. A fast SHAKE algorithm to solve distance constraint equations for small molecules in molecular dynamics simulations. J. Comput. Chem. 2001, 22, 501–508. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef] [Green Version]
- Bakan, A.; Meireles, L.M.; Bahar, I. ProDy: Protein dynamics inferred from theory and experiments. Bioinformatics 2011, 27, 1575–1577. [Google Scholar] [CrossRef] [Green Version]
Sample Availability: Samples of the compounds are not available from the authors. |
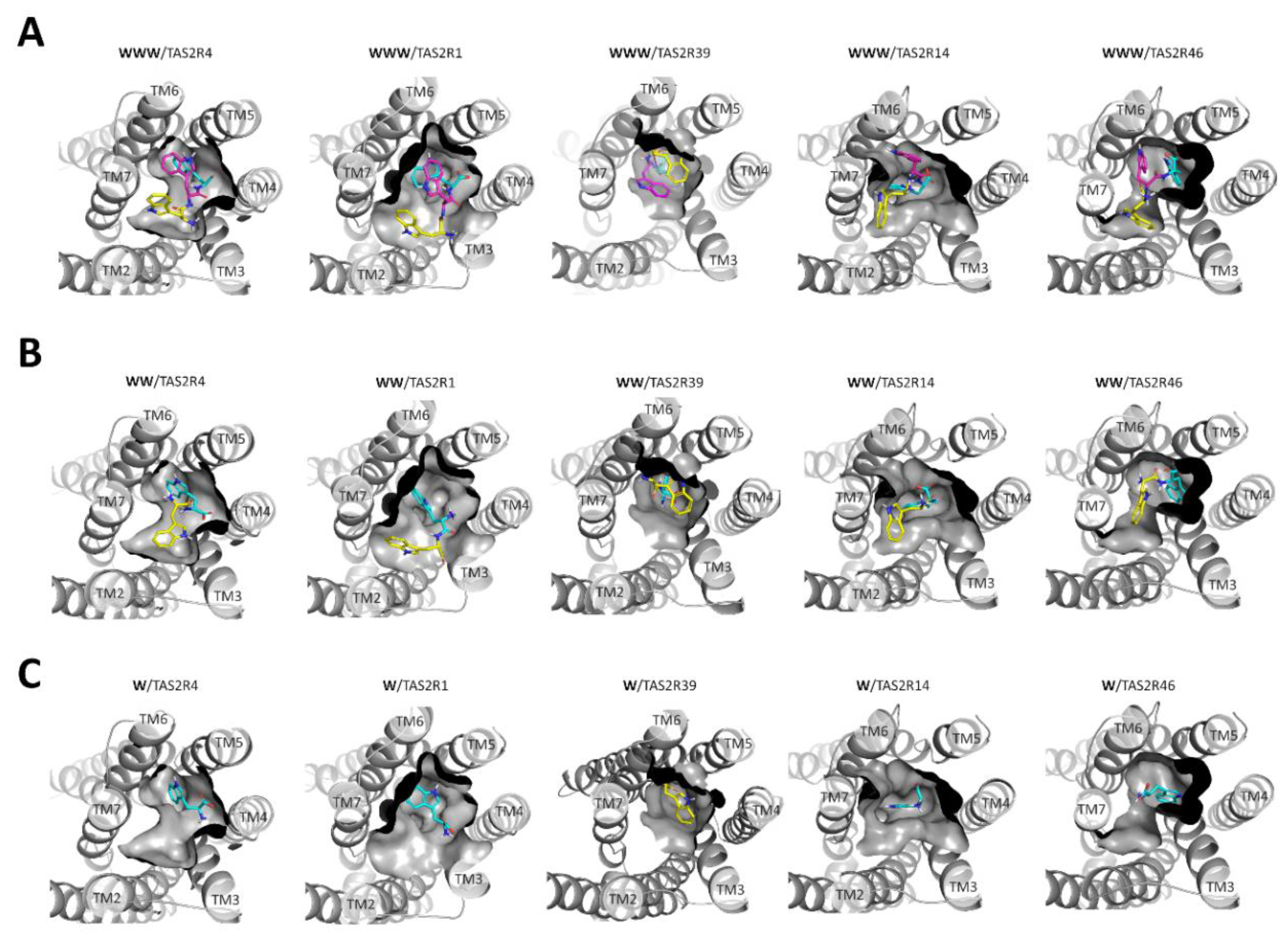
| TAS2R4 | TAS2R1 | TAS2R39 | TAS2R14 | TAS2R46 | ||
|---|---|---|---|---|---|---|
| WWW | Activity (mM) | 0.01 (EC50: 0.03 ± 0.005) | 0.1 | 0.1 | 0.1 | 0.1 |
| Glide score | −10.88 | −8.82 | −8.10 | −8.48 | −8.20 | |
| WW | Activity (mM) | 1.0 (EC50: 0.66 ± 0.03) | 0.3 | 1.0 | ||
| Glide score | −8.59 | −8.71 | −8.35 | −5.55 | −5.27 | |
| W | Activity (mM) | 10 | ||||
| Glide score | −7.05 | −5.36 | −5.34 | −5.43 | −5.05 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Pizio, A.; Nicoli, A. In Silico Molecular Study of Tryptophan Bitterness. Molecules 2020, 25, 4623. https://doi.org/10.3390/molecules25204623
Di Pizio A, Nicoli A. In Silico Molecular Study of Tryptophan Bitterness. Molecules. 2020; 25(20):4623. https://doi.org/10.3390/molecules25204623
Chicago/Turabian StyleDi Pizio, Antonella, and Alessandro Nicoli. 2020. "In Silico Molecular Study of Tryptophan Bitterness" Molecules 25, no. 20: 4623. https://doi.org/10.3390/molecules25204623