Misprediction of Structural Disorder in Halophiles
Abstract
:1. Introduction
2. Results
2.1. Species Distribution of the DisProt Database
2.2. Assessing Structural Disorder in Extremophile Groups
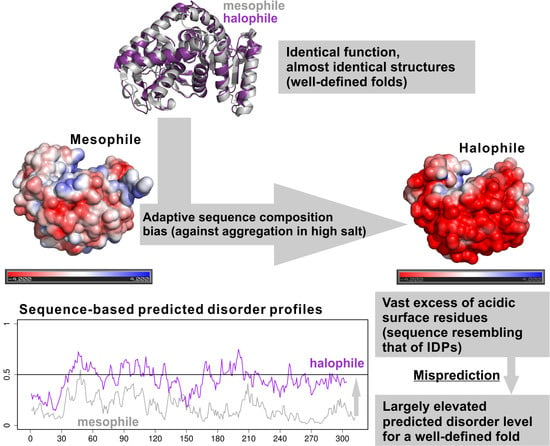
2.3. Halophile–Mesophile Protein Homologs with Available Structures Clearly Support Overprediction of Disorder in Halophiles
2.4. Archaea have Lower Disorder Levels than Bacteria due to Their Bias Towards Hyperthermophilic Species
3. Discussion
4. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Habchi, J.; Tompa, P.; Longhi, S.; Uversky, V.N. Introducing protein intrinsic disorder. Chem. Rev. 2014, 114, 6561–6588. [Google Scholar] [CrossRef] [PubMed]
- Van der Lee, R.; Buljan, M.; Lang, B.; Weatheritt, R.J.; Daughdrill, G.W.; Dunker, A.K.; Fuxreiter, M.; Gough, J.; Gsponer, J.; Jones, D.T.; et al. Classification of intrinsically disordered regions and proteins. Chem. Rev. 2014, 114, 6589–6631. [Google Scholar] [CrossRef] [PubMed]
- Wright, P.E.; Dyson, H.J. Intrinsically unstructured proteins: Re-assessing the protein structure-function paradigm. J. Mol. Biol. 1999, 293, 321–331. [Google Scholar] [CrossRef] [PubMed]
- Wright, P.E.; Dyson, H.J. Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 2015, 16, 18–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pancsa, R.; Tompa, P. Structural disorder in eukaryotes. PloS ONE 2012, 7, e34687. [Google Scholar] [CrossRef] [PubMed]
- Ward, J.J.; Sodhi, J.S.; McGuffin, L.J.; Buxton, B.F.; Jones, D.T. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J. Mol. Biol. 2004, 337, 635–645. [Google Scholar] [CrossRef]
- Sickmeier, M.; Hamilton, J.A.; LeGall, T.; Vacic, V.; Cortese, M.S.; Tantos, A.; Szabo, B.; Tompa, P.; Chen, J.; Uversky, V.N.; et al. DisProt: The Database of Disordered Proteins. Nucleic Acids Res. 2007, 35, D786–D793. [Google Scholar] [CrossRef]
- Dosztanyi, Z.; Meszaros, B.; Simon, I. Bioinformatical approaches to characterize intrinsically disordered/unstructured proteins. Briefings Bioinf. 2010, 11, 225–243. [Google Scholar] [CrossRef]
- Ferron, F.; Longhi, S.; Canard, B.; Karlin, D. A practical overview of protein disorder prediction methods. Proteins 2006, 65, 1–14. [Google Scholar] [CrossRef]
- Cambillau, C.; Claverie, J.M. Structural and genomic correlates of hyperthermostability. J. Biol. Chem. 2000, 275, 32383–32386. [Google Scholar] [CrossRef]
- Imanaka, T. Molecular bases of thermophily in hyperthermophiles. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2011, 87, 587–602. [Google Scholar] [CrossRef] [Green Version]
- Kiraga, J.; Mackiewicz, P.; Mackiewicz, D.; Kowalczuk, M.; Biecek, P.; Polak, N.; Smolarczyk, K.; Dudek, M.R.; Cebrat, S. The relationships between the isoelectric point and: Length of proteins, taxonomy and ecology of organisms. BMC Genom. 2007, 8, 163. [Google Scholar] [CrossRef] [PubMed]
- Knight, C.G.; Kassen, R.; Hebestreit, H.; Rainey, P.B. Global analysis of predicted proteomes: Functional adaptation of physical properties. Proc. Natl. Acad. Sci. USA 2004, 101, 8390–8395. [Google Scholar] [CrossRef] [PubMed]
- Li, W.F.; Zhou, X.X.; Lu, P. Structural features of thermozymes. Biotechnol. Adv. 2005, 23, 271–281. [Google Scholar] [CrossRef]
- Pollo, S.M.; Zhaxybayeva, O.; Nesbo, C.L. Insights into thermoadaptation and the evolution of mesophily from the bacterial phylum Thermotogae. Can. J. Microbiol. 2015, 61, 655–670. [Google Scholar] [CrossRef] [Green Version]
- Robb, F.T.; Clark, D.S. Adaptation of proteins from hyperthermophiles to high pressure and high temperature. J. Mol. Microbiol. Biotechnol. 1999, 1, 101–105. [Google Scholar]
- Szilagyi, A.; Zavodszky, P. Structural differences between mesophilic, moderately thermophilic and extremely thermophilic protein subunits: Results of a comprehensive survey. Structure 2000, 8, 493–504. [Google Scholar] [CrossRef]
- Taylor, T.J.; Vaisman, II. Discrimination of thermophilic and mesophilic proteins. BMC Struct. Biol. 2010, 10 (Suppl. 1), S5. [Google Scholar] [CrossRef] [Green Version]
- Meszaros, B.; Simon, I.; Dosztanyi, Z. Prediction of protein binding regions in disordered proteins. PLoS Comput. Biol. 2009, 5, e1000376. [Google Scholar] [CrossRef]
- Vicedo, E.; Schlessinger, A.; Rost, B. Environmental Pressure May Change the Composition Protein Disorder in Prokaryotes. PloS ONE 2015, 10, e0133990. [Google Scholar] [CrossRef] [PubMed]
- Xue, B.; Dunker, A.K.; Uversky, V.N. Orderly order in protein intrinsic disorder distribution: Disorder in 3500 proteomes from viruses and the three domains of life. J. Biomol. Struct. Dyn. 2012, 30, 137–149. [Google Scholar] [CrossRef] [PubMed]
- Xue, B.; Williams, R.W.; Oldfield, C.J.; Dunker, A.K.; Uversky, V.N. Archaic chaos: Intrinsically disordered proteins in Archaea. BMC Syst. Biol. 2010, 4 (Suppl. 1), S1. [Google Scholar] [CrossRef]
- Dunker, A.K.; Lawson, J.D.; Brown, C.J.; Williams, R.M.; Romero, P.; Oh, J.S.; Oldfield, C.J.; Campen, A.M.; Ratliff, C.M.; Hipps, K.W.; et al. Intrinsically disordered protein. J. Mol. Graphics Modell. 2001, 19, 26–59. [Google Scholar] [CrossRef] [Green Version]
- Tompa, P. Intrinsically unstructured proteins. Trends Biochem. Sci. 2002, 27, 527–533. [Google Scholar] [CrossRef]
- Uversky, V.N.; Gillespie, J.R.; Fink, A.L. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins 2000, 41, 415–427. [Google Scholar] [CrossRef]
- Elcock, A.H.; McCammon, J.A. Electrostatic contributions to the stability of halophilic proteins. J. Mol. Biol. 1998, 280, 731–748. [Google Scholar] [CrossRef] [PubMed]
- Graziano, G.; Merlino, A. Molecular bases of protein halotolerance. Biochim. Biophys. Acta 2014, 1844, 850–858. [Google Scholar] [CrossRef] [PubMed]
- The UniProt, C. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2017, 45, D158–D169. [Google Scholar] [CrossRef]
- Calligari, P.A.; Calandrini, V.; Ollivier, J.; Artero, J.B.; Hartlein, M.; Johnson, M.; Kneller, G.R. Adaptation of Extremophilic Proteins with Temperature and Pressure: Evidence from Initiation Factor 6. J. Phys. Chem. B 2015, 119, 7860–7873. [Google Scholar] [CrossRef] [PubMed]
- Katava, M.; Kalimeri, M.; Stirnemann, G.; Sterpone, F. Stability and Function at High Temperature. What Makes a Thermophilic GTPase Different from Its Mesophilic Homologue. J. Phys. Chem. B 2016, 120, 2721–2730. [Google Scholar] [CrossRef] [PubMed]
- Burra, P.V.; Kalmar, L.; Tompa, P. Reduction in structural disorder and functional complexity in the thermal adaptation of prokaryotes. PloS ONE 2010, 5, e12069. [Google Scholar] [CrossRef] [PubMed]
- Pancsa, R.; Tompa, P. Essential functions linked with structural disorder in organisms of minimal genome. Biol. Direct 2016, 11, 45. [Google Scholar] [CrossRef] [PubMed]
- Litchfield, C.D. Halophiles. J. Ind. Microbiol. Biotechnol. 2002, 28, 21–22. [Google Scholar] [CrossRef]
- Krisko, A.; Smole, Z.; Debret, G.; Nikolic, N.; Radman, M. Unstructured hydrophilic sequences in prokaryotic proteomes correlate with dehydration tolerance and host association. J. Mol. Biol. 2010, 402, 775–782. [Google Scholar] [CrossRef] [PubMed]
- Boothby, T.C.; Pielak, G.J. Intrinsically Disordered Proteins and Desiccation Tolerance: Elucidating Functional and Mechanistic Underpinnings of Anhydrobiosis. BioEssays 2017, 39. [Google Scholar] [CrossRef] [PubMed]
- Tompa, P.; Csermely, P. The role of structural disorder in the function of RNA and protein chaperones. FASEB J. 2004, 18, 1169–1175. [Google Scholar] [CrossRef] [PubMed]
- Hundertmark, M.; Hincha, D.K. LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genom. 2008, 9, 118. [Google Scholar] [CrossRef]
- Boothby, T.C.; Tapia, H.; Brozena, A.H.; Piszkiewicz, S.; Smith, A.E.; Giovannini, I.; Rebecchi, L.; Pielak, G.J.; Koshland, D.; Goldstein, B. Tardigrades Use Intrinsically Disordered Proteins to Survive Desiccation. Mol. Cell. 2017, 65, 975–984. [Google Scholar] [CrossRef]
- Dosztanyi, Z.; Csizmok, V.; Tompa, P.; Simon, I. The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J. Mol. Biol. 2005, 347, 827–839. [Google Scholar] [CrossRef]
- Prilusky, J.; Felder, C.E.; Zeev-Ben-Mordehai, T.; Rydberg, E.H.; Man, O.; Beckmann, J.S.; Silman, I.; Sussman, J.L. FoldIndex: A simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics 2005, 21, 3435–3438. [Google Scholar] [CrossRef]
- Yang, Z.R.; Thomson, R.; McNeil, P.; Esnouf, R.M. RONN: The bio-basis function neural network technique applied to the detection of natively disordered regions in proteins. Bioinformatics 2005, 21, 3369–3376. [Google Scholar] [CrossRef] [PubMed]
- Obradovic, Z.; Peng, K.; Vucetic, S.; Radivojac, P.; Dunker, A.K. Exploiting heterogeneous sequence properties improves prediction of protein disorder. Proteins 2005, 61 (Suppl. 7), 176–182. [Google Scholar] [CrossRef]
- Ishida, T.; Kinoshita, K. PrDOS: Prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 2007, 35, W460–W464. [Google Scholar] [CrossRef] [PubMed]
- Hecker, J.; Yang, J.Y.; Cheng, J. Protein disorder prediction at multiple levels of sensitivity and specificity. BMC Genom. 2008, 9 (Suppl. 1), S9. [Google Scholar] [CrossRef] [Green Version]
- Linding, R.; Russell, R.B.; Neduva, V.; Gibson, T.J. GlobPlot: Exploring protein sequences for globularity and disorder. Nucleic Acids Res. 2003, 31, 3701–3708. [Google Scholar] [CrossRef] [PubMed]
- Walsh, I.; Martin, A.J.; Di Domenico, T.; Vullo, A.; Pollastri, G.; Tosatto, S.C. CSpritz: Accurate prediction of protein disorder segments with annotation for homology, secondary structure and linear motifs. Nucleic Acids Res. 2011, 39, W190–W196. [Google Scholar] [CrossRef]
- Bohm, G.; Jaenicke, R. A structure-based model for the halophilic adaptation of dihydrofolate reductase from Halobacterium volcanii. Protein Eng. 1994, 7, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Pieper, U.; Kapadia, G.; Mevarech, M.; Herzberg, O. Structural features of halophilicity derived from the crystal structure of dihydrofolate reductase from the Dead Sea halophilic archaeon, Haloferax volcanii. Structure 1998, 6, 75–88. [Google Scholar] [CrossRef] [Green Version]
- Dym, O.; Mevarech, M.; Sussman, J.L. Structural features that stabilize halophilic malate dehydrogenase from an archaebacterium. Science 1995, 267, 1344–1346. [Google Scholar] [CrossRef]
- Frolow, F.; Harel, M.; Sussman, J.L.; Mevarech, M.; Shoham, M. Insights into protein adaptation to a saturated salt environment from the crystal structure of a halophilic 2Fe-2S ferredoxin. Nat. Struct. Biol. 1996, 3, 452–458. [Google Scholar] [CrossRef]
- Winter, J.A.; Christofi, P.; Morroll, S.; Bunting, K.A. The crystal structure of Haloferax volcanii proliferating cell nuclear antigen reveals unique surface charge characteristics due to halophilic adaptation. BMC Struct. Biol. 2009, 9, 55. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.; Liu, M.Y.; Chang, T.; Li, M.; Le Gall, J.; Gui, L.L.; Zhang, J.P.; Jiang, T.; Liang, D.C.; Chang, W.R. Three-dimensional structure of manganese superoxide dismutase from Bacillus halodenitrificans, a component of the so-called “green protein”. J. Struct. Biol. 2002, 139, 171–180. [Google Scholar] [CrossRef]
- Baldacci, G.; Guinet, F.; Tillit, J.; Zaccai, G.; de Recondo, A.M. Functional implications related to the gene structure of the elongation factor EF-Tu from Halobacterium marismortui. Nucleic Acids Res. 1990, 18, 507–511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Britton, K.L.; Stillman, T.J.; Yip, K.S.; Forterre, P.; Engel, P.C.; Rice, D.W. Insights into the molecular basis of salt tolerance from the study of glutamate dehydrogenase from Halobacterium salinarum. J. Biol. Chem. 1998, 273, 9023–9030. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Uversky, V.N.; Kurgan, L. Comprehensive review of methods for prediction of intrinsic disorder and its molecular functions. Cell. Mol. Life Sci. 2017, 74, 3069–3090. [Google Scholar] [CrossRef] [PubMed]
- Arakawa, T.; Yamaguchi, R.; Tokunaga, H.; Tokunaga, M. Unique Features of Halophilic Proteins. Curr. Protein Pept. Sci. 2017, 18, 65–71. [Google Scholar] [CrossRef]
- Lenton, S.; Walsh, D.L.; Rhys, N.H.; Soper, A.K.; Dougan, L. Structural evidence for solvent-stabilisation by aspartic acid as a mechanism for halophilic protein stability in high salt concentrations. Phys. Chem. Chem. Phys. 2016, 18, 18054–18062. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Natale, D.A.; Finn, R.D.; Huang, H.; Zhang, J.; Wu, C.H.; Mazumder, R. Representative proteomes: A stable, scalable and unbiased proteome set for sequence analysis and functional annotation. PloS ONE 2011, 6, e18910. [Google Scholar] [CrossRef]
- Pagani, I.; Liolios, K.; Jansson, J.; Chen, I.M.; Smirnova, T.; Nosrat, B.; Markowitz, V.M.; Kyrpides, N.C. The Genomes OnLine Database (GOLD) v.4: Status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 2012, 40, D571–D579. [Google Scholar] [CrossRef]






| Mesophile Species | Hyperthermophile Species | Number of 1:1 Protein Orthologs | Number of LDRs (≥40 res.) in Mesophile | Number of LDRs with ≥75% of Length Preserved (of which Ribosomal) |
|---|---|---|---|---|
| Clostridium perfringens (ATCC 13124) | Caldicellulosiruptor saccharolyticus (ATCC 43494) | 717 | 15 | 6 (3) |
| Clostridium perfringens (ATCC 13124) | Thermaerobacter marianensis (ATCC 700841) | 539 | 9 | 7 (3) |
| Clostridium perfringens (ATCC 13124) | Thermoanaerobacter tengcongensis (DSM 15242) | 782 | 15 | 7 (3) |
| Desulfitobacterium hafniense (DSM 10664) | Carboxydothermus hydrogenoformans (DSM 6008) | 1109 | 36 | 15 (3) |
| Methanococcus maripaludis (S2) | Methanocaldococcus jannaschii (DSM 2661) | 1078 | 8 | 6 (4) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pancsa, R.; Kovacs, D.; Tompa, P. Misprediction of Structural Disorder in Halophiles. Molecules 2019, 24, 479. https://doi.org/10.3390/molecules24030479
Pancsa R, Kovacs D, Tompa P. Misprediction of Structural Disorder in Halophiles. Molecules. 2019; 24(3):479. https://doi.org/10.3390/molecules24030479
Chicago/Turabian StylePancsa, Rita, Denes Kovacs, and Peter Tompa. 2019. "Misprediction of Structural Disorder in Halophiles" Molecules 24, no. 3: 479. https://doi.org/10.3390/molecules24030479