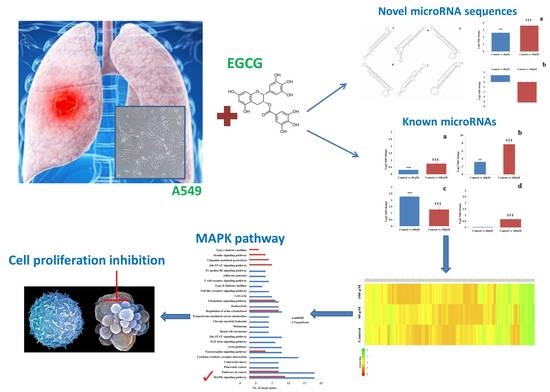
Next-Generation Sequencing Reveals the Role of Epigallocatechin-3-Gallate in Regulating Putative Novel and Known microRNAs Which Target the MAPK Pathway in Non-Small-Cell Lung Cancer A549 Cells
Abstract
:1. Introduction
2. Results
2.1. EGCG Induced G0/G1 Phase Arrest in A549 Cell Line
2.2. Analysis of MicroRNAs
2.3. A549 Cell Associated MicroRNA Expression Signature
2.4. Prediction of Putative Novel MicroRNAs
2.5. Differential Expression Analysis of Known MicroRNAs
2.6. Differential Expression Analysis of Putative Novel MicroRNA Sequences
2.7. qRT-PCR Analysis of MicroRNAs
2.8. KEGG and PANTHER Pathway Enrichment of Targets of Validated microRNAs
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Treatment
4.2. Next-Generation Sequencing
4.3. Classification and Differential Expression Analysis of MicroRNAs
4.4. Validation of MicroRNAs
4.5. KEGG and PANTHER Pathway Enrichment of Targets of Validated MicroRNAs
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Min, K.; Kwon, T.K. Anticancer effects and molecular mechanisms of epigallocatechin-3-gallate. Integr. Med. Res. 2014, 3, 16–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, T.; Katiyar, S.K. Green tea polyphenol, (−)-epigallocatechin-3-gallate, induces toxicity in human skin cancer cells by targeting beta-catenin signaling. Toxicol. Appl. Pharmacol. 2013, 273, 418–424. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.-H.; Wang, X.; Feng, Q. EGCG enhances the efficacy of cisplatin by downregulating hsa-miR-98-5p in NSCLC A549 cells. Nutr. Cancer 2014, 66, 636–644. [Google Scholar] [CrossRef] [PubMed]
- Jankun, J.; Selman, S.H.; Swiercz, R.; Skrzypczak-Jankun, E. Why drinking green tea could prevent cancer. Nature 1997, 387, 561. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, M.; Deguchi, A.; Lim, J.T.E.; Moriwaki, H.; Kopelovich, L.; Weinstein, I.B. (−)-Epigallocatechin gallate and polyphenon E inhibit growth and activation of the epidermal growth factor receptor and human epidermal growth factor receptor-2 signaling pathways in human colon cancer cells. Clin. Cancer Res. 2005, 11, 2735–2746. [Google Scholar] [CrossRef] [PubMed]
- García, V.; Lara-chica, M.; Cantarero, I.; Sterner, O.; Marco, A.; Muñoz, E. Galiellalactone induces cell cycle arrest and apoptosis through the ATM/ATR pathway in prostate cancer cells. Oncotarget 2015, 7. [Google Scholar] [CrossRef] [PubMed]
- Mukhtar, H.; Ahmad, N. Tea polyphenols: Prevention of cancer and optimizing health. Am. J. Clin. Nutr. 2000, 71, 1698S–1702S, discussion 1703S–1704S. [Google Scholar] [CrossRef]
- Yang, C.S.; Maliakal, P.; Meng, X. Inhibition of carcinogenesis by tea. Annu. Rev. Pharmacol. Toxicol. 2002, 42, 25–54. [Google Scholar] [CrossRef]
- Jemal, A.; Siegel, R.; Ward, E.; Hao, Y.; Xu, J.; Thun, M.J. Cancer Statistics. CA Cancer J. Clin. 2009, 59, 225–249. [Google Scholar] [CrossRef]
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-Tieulent, J.; Jemal, A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015, 65, 87–108. [Google Scholar] [CrossRef]
- Molina, J.R.; Yang, P.; Cassivi, S.D.; Schild, S.E.; Adjei, A.A. Non-small cell lung cancer: Epidemiology, risk factors, treatment, and survivorship. Mayo Clin. Proc. 2008, 83, 584–594. [Google Scholar] [CrossRef]
- Sun, S.; Schiller, J.H.; Gazdar, A.F. Lung cancer in never smokers—A different disease. Nat. Rev. Cancer 2007, 7, 778–790. [Google Scholar] [CrossRef] [PubMed]
- Condoluci, A.; Mazzara, C.; Zoccoli, A.; Pezzuto, A.; Tonini, G. Impact of smoking on lung cancer treatment effectiveness: A review. Future Oncol. 2016, 12, 2149–2161. [Google Scholar] [CrossRef] [PubMed]
- Sini, C.; Tuzi, A.; Rossi, G.; Russo, A.; Pezzuto, A. Acquired resistance in oncogene-addicted non-small-cell lung cancer. Future Oncol. 2018, 14, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Pezzuto, A.; Manicone, M.; Scaini, M.C.; Ricci, A.; Mariotta, S.; Zamarchi, R.; Rossi, E. What information could the main actors of liquid biopsy provide?—A representative case of non-small cell lung cancer (NSCLC). J. Thorac. Dis. 2018, 10, E570–E576. [Google Scholar] [CrossRef] [PubMed]
- Stahlhut, C.; Slack, F.J. MicroRNAs and the cancer phenotype: Profiling, signatures and clinical implications. Genome Med. 2013, 5, 111. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, Y.; Tay, F.C.; Lam, D.H.; Sandanaraj, E.; Tang, C.; Ang, B.T.; Wang, S. Attenuated adenosine-to-inosine editing of microRNA-376a * promotes invasiveness of glioblastoma cells. J. Clin. Investig. 2012, 122, 4059–4076. [Google Scholar] [CrossRef]
- Dhillon, A.S.; Hagan, S.; Rath, O.; Kolch, W. MAP kinase signalling pathways in cancer. Oncogene 2007, 26, 3279–3290. [Google Scholar] [CrossRef] [Green Version]
- Sarris, E.G.; Saif, M.W.; Syrigos, K.N. The biological role of PI3K pathway in lung cancer. Pharmaceuticals 2012, 5, 1236–1264. [Google Scholar] [CrossRef]
- Thorpe, L.M.; Yuzugullu, H.; Zhao, J.J. PI3K in cancer: Divergent roles of isoforms, modes of activation and therapeutic targeting. Nat. Rev. Cancer 2014, 15, 7. [Google Scholar] [CrossRef]
- Thomas, S.J.; Snowden, J.A.; Zeidler, M.P.; Danson, S.J. The role of JAK/STAT signalling in the pathogenesis, prognosis and treatment of solid tumours. Br. J. Cancer 2015, 113, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Zhan, T.; Rindtorff, N.; Boutros, M. Wnt signaling in cancer. Oncogene 2017, 36, 1461–1473. [Google Scholar] [CrossRef]
- De Luca, A.; Maiello, M.R.; D’Alessio, A.; Pergameno, M.; Normanno, N. The RAS/RAF/MEK/ERK and the PI3K/AKT signalling pathways: Role in cancer pathogenesis and implications for therapeutic approaches. Expert Opin. Ther. Targets 2012, 16 (Suppl. 2), S17–S27. [Google Scholar] [CrossRef] [PubMed]
- McCubrey, J.A.; Steelman, L.S.; Chappell, W.H.; Abrams, S.L.; Wong, E.W.T.; Chang, F.; Lehmann, B.; Terrian, D.M.; Milella, M.; Tafuri, A.; et al. Roles of the Raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance. Biochim. Biophys. Acta Mol. Cell Res. 2007, 1773, 1263–1284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, M.A.; Wappett, M.; Delpuech, O.; Brown, H.; Chresta, C.M. Enhanced MAPK signaling drives ETS1-mediated induction of miR-29b leading to downregulation of TET1 and changes in epigenetic modifications in a subset of lung SCC. Oncogene 2016, 35, 4345–4357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, L.; Middleton, J.; Jeon, Y.J.; Magee, P.; Veneziano, D.; Laganà, A.; Leong, H.S.; Sahoo, S.; Fassan, M.; Booton, R.; et al. KRAS induces lung tumorigenesis through microRNAs modulation article. Cell Death Dis. 2018, 9. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597. [Google Scholar] [CrossRef]
- Friedman, R.C.; Farh, K.K.-H.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef]
- Calin, G.A.; Sevignani, C.; Dumitru, C.D.; Hyslop, T.; Noch, E.; Yendamuri, S.; Shimizu, M.; Rattan, S.; Bullrich, F.; Negrini, M.; et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. USA 2004, 101, 2999–3004. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Jennings, S.F.; Tong, W.; Hong, H. Next generation sequencing for profiling expression of miRNAs: Technical progress and applications in drug development. J. Biomed. Sci. Eng. 2011, 4, 666–676. [Google Scholar] [CrossRef] [PubMed]
- Duncavage, E.; Goodgame, B.; Sezhiyan, A.; Govindan, R.; Pfeifer, J. Use of MicroRNA expression levels to predict outcomes in resected stage i non-small cell lung cancer. J. Thorac. Oncol. 2010, 5, 1755–1763. [Google Scholar] [CrossRef] [PubMed]
- Markou, A.; Sourvinou, I.; Vorkas, P.A.; Yousef, G.M.; Lianidou, E. Clinical evaluation of microRNA expression profiling in non small cell lung cancer. Lung Cancer 2013, 81, 388–396. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, J.; Lin, Z.; Cao, J.; Huang, H.; Jiang, Y.; He, H.; Yang, L.; Ren, N.; Liu, G. Role of deregulated microRNAs in non-small cell lung cancer progression using fresh-frozen and formalin-fixed, paraffin-embedded samples. Oncol. Lett. 2016, 11, 801–808. [Google Scholar] [CrossRef] [PubMed]
- Gallach, S.; Calabuig-Fariñas, S.; Jantus-Lewintre, E.; Camps, C. MicroRNAs: Promising new antiangiogenic targets in cancer. Biomed. Res. Int. 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- Ji, Z.; Wang, G.; Zhang, C.; Xie, Z.; Liu, Z.; Wang, J. Identification and function prediction of novel MicroRNAs in Laoshan dairy goats. Asian-Australas. J. Anim. Sci. 2013, 26, 309–315. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.-C.; Li, C.; Gao, F.; Xu, Y.; Jiang, Z.-B.; Liu, J.-X.; Jin, L.-Y. Epigallocatechin gallate inhibits the growth of human lung cancer by directly targeting the EGFR signaling pathway. Oncol. Rep. 2014, 31, 1343–1349. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Hussain, T.; Mukhtar, H. Molecular pathway for (−)-epigallocatechin-3-gallate-induced cell cycle arrest and apoptosis of human prostate carcinoma cells. Arch. Biochem. Biophys. 2003, 410, 177–185. [Google Scholar] [CrossRef]
- Masuda, M.; Suzui, M.; Weinstein, I.B. Effects of epigallocatechin-3-gallate on growth, epidermal growth factor receptor signaling pathways, gene expression, and chemosensitivity in human head and neck squamous cell carcinoma cell lines. Clin. Cancer Res. 2001, 7, 4220–4229. [Google Scholar]
- Ahmad, N.; Cheng, P.; Mukhtar, H. Cell cycle dysregulation by green tea polyphenol epigallocatechin-3-gallate. Biochem. Biophys. Res. Commun. 2000, 275, 328–334. [Google Scholar] [CrossRef]
- Kweon, M.-H.; Saleem, M.; Mukhtar, H. Constitutive overexpression of Nrf2-dependent HO-1 confers resistance to apoptosis by EGCG in human lung adenocarcinoma A549 cells. Cancer Res. 2006, 66, 1149. [Google Scholar]
- Hou, Z.; Sang, S.; You, H.; Lee, M.J.; Hong, J.; Chin, K.V.; Yang, C.S. Mechanism of action of (−)-epigallocatechin-3-gallate: Auto-oxidation- dependent inactivation of epidermal growth factor receptor and direct effects on growth inhibition in human esophageal cancer KYSE 150 cells. Cancer Res. 2005, 65, 8049–8056. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.; Lu, H.; Meng, X.; Colon, H.-H.; Ryu, J.; Hara, Y.; Yang, C.S. Stability, Cellular Uptake, Biotransformation, and Efflux of Tea Polyphenol (−)-Epigallocatechin-3-Gallate in HT-29 Human Colon Adenocarcinoma Cells Stability, Cellular Uptake, Biotransformation, and Efflux of Tea Polyphenol. Cancer Res. 2002, 62, 7241–7246. [Google Scholar]
- Xue, X.; Liu, Y.; Wang, Y.; Meng, M.; Wang, K.; Zang, X.; Zhao, S.; Sun, X.; Cui, L.; Pan, L.; et al. MiR-21 and MiR-155 promote non-small cell lung cancer progression by downregulating SOCS1, SOCS6, and PTEN. Oncotarget 2016, 7, 84508–84519. [Google Scholar] [CrossRef] [Green Version]
- Markou, A.; Zavridou, M.; Lianidou, E.S. MiRNA-21 as a novel therapeutic target in lung cancer. Lung Cancer Targets Ther. 2016, 7, 19–27. [Google Scholar] [CrossRef]
- Li, C.; Yin, Y.; Liu, X.; Xi, X.; Xue, W.; Qu, Y. Non-small cell lung cancer associated microRNA expression signature: Integrated bioinformatics analysis, validation and clinical significance. Oncotarget 2017, 8, 24564–24578. [Google Scholar] [CrossRef]
- Medina, P.P.; Nolde, M.; Slack, F.J. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature 2010, 467, 86–90. [Google Scholar] [CrossRef]
- Volinia, S.; Calin, G.A.; Liu, C.-G.; Ambs, S.; Cimmino, A.; Petrocca, F.; Visone, R.; Iorio, M.; Roldo, C.; Ferracin, M.; et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 2006, 103, 2257–2261. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Meng, H.; Peng, Q.; Yang, X.; Gan, R.; Zhao, L.; Chen, Z.; Lu, J.; Meng, Q.H. Downregulation of microRNA-21 expression restrains non-small cell lung cancer cell proliferation and migration through upregulation of programmed cell death 4. Cancer Gene Ther. 2015, 22, 23–29. [Google Scholar] [CrossRef]
- Buscaglia, L.E.B.; Li, Y. Apoptosis and the target genes of microRNA-21. Chin. J. Cancer 2011, 30, 371–380. [Google Scholar] [CrossRef] [Green Version]
- Fujita, Y.; Yagishita, S.; Hagiwara, K.; Yoshioka, Y.; Kosaka, N.; Takeshita, F.; Fujiwara, T.; Tsuta, K.; Nokihara, H.; Tamura, T.; et al. The clinical relevance of the miR-197/CKS1B/STAT3-mediated PD-L1 network in chemoresistant non-small-cell lung cancer. Mol. Ther. 2015, 23, 717–727. [Google Scholar] [CrossRef] [PubMed]
- Fassina, A.; Cappellesso, R.; Fassan, M. Classification of non-small cell lung carcinoma in transthoracic needle specimens using microRNA expression profiling. Chest 2011, 140, 1305–1311. [Google Scholar] [CrossRef] [PubMed]
- Yanaihara, N.; Caplen, N.; Bowman, E.; Seike, M.; Kumamoto, K.; Yi, M.; Stephens, R.M.; Okamoto, A.; Yokota, J.; Tanaka, T.; et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 2006, 9, 189–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, L.; Huang, Q.; Zhang, S.; Zhang, Q.; Chang, J.; Qiu, X.; Wang, E. Hsa-miR-125a-3p and hsa-miR-125a-5p are downregulated in non-small cell lung cancer and have inverse effects on invasion and migration of lung cancer cells. BMC Cancer 2010, 10. [Google Scholar] [CrossRef] [PubMed]
- Liang, T.; Guo, L.; Liu, C. Genome-wide analysis of mir-548 gene family reveals evolutionary and functional implications. J. Biomed. Biotechnol. 2012, 2012. [Google Scholar] [CrossRef] [PubMed]
- Rane, J.K.; Scaravilli, M.; Ylipää, A.; Pellacani, D.; Mann, V.M.; Simms, M.S.; Nykter, M.; Collins, A.T.; Visakorpi, T.; Maitland, N.J. MicroRNA expression profile of primary prostate cancer stem cells as a source of biomarkers and therapeutic targets. Eur. Urol. 2015, 67, 7–10. [Google Scholar] [CrossRef] [PubMed]
- Incoronato, M.; Garofalo, M.; Urso, L.; Romano, G.; Quintavalle, C.; Zanca, C.; Iaboni, M.; Nuovo, G.; Croce, C.M.; Condorelli, G. miR-212 increases tumor necrosis factor-related apoptosis-inducing ligand sensitivity in non-small cell lung cancer by targeting the antiapoptotic protein PED. Cancer Res. 2010, 70, 3638–3646. [Google Scholar] [CrossRef]
- Jiang, X.; Chen, X.; Chen, L.; Ma, Y.; Zhou, L.; Qi, Q.; Liu, Y.; Zhang, S.; Luo, J.; Zhou, X. Upregulation of the miR-212/132 cluster suppresses proliferation of human lung cancer cells. Oncol. Rep. 2015, 33, 705–712. [Google Scholar] [CrossRef]
- Xiang, W.; Wu, X.; Huang, C.; Wang, M.; Zhao, X.; Luo, G. PTTG1 regulated by miR-146a-3p promotes bladder cancer migration, invasion, metastasis and growth. Oncotarget 2016, 8, 1–15. [Google Scholar] [CrossRef]
- Patnaik, S.K.; Kannisto, E.; Mallick, R.; Yendamuri, S. Overexpression of the lung cancer-prognostic miR-146b microRNAs has a minimal and negative effect on the malignant phenotype of A549 lung cancer cells. PLoS ONE 2011, 6. [Google Scholar] [CrossRef]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucl. Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Riffo-Campos, Á.L.; Riquelme, I.; Brebi-Mieville, P. Tools for sequence-based miRNA target prediction: What to choose? Int. J. Mol. Sci. 2016, 17, 1987. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |













| Putative Novel MicroRNA ID | Sequences | Chromosome Number | Precursor Start | Precursor End | Mature Start | Mature End | Precursor Sequence | MFE (Kcal/mol) |
|---|---|---|---|---|---|---|---|---|
| A549 Control | ||||||||
| TPP-A549-1 | CCAGGAUGCACGCUCGCUGGGCU | 17 | 375,2068 | 375,2154 | 375,2078 | 375,2100 | CCGUUGCCACCCAGGAUGCACGCUCGCUGGGCUGGUGGGUGCCCACUCCGUUGCCACCCAGGAUGCACGCUCACUGGCCAGGGACCU | −30.7 |
| TPP−A549-2 | CGCUUCGUGUAGACCCUCCAC | 15 | 748,733,11 | 748,733,96 | 748,733,21 | 748,733,41 | GACCCAGCGGCGCUUCGUGUAGACCCUCCACUUCCGGGAGCGAGGCAGCGGUUCUGGCGCAGGCGCGAUGCCCUCCCCCGAGGGCG | −35.4 |
| TPP-A549-3 | CUUAGAAUUCCGAUGCUGGGG | 22 | 267,618,39 | 267,619,28 | 267,618,98 | 267,619,18 | AGAUUCUGGACUUAGAAUUCCGAUGCUGGGGCCAGGCACAGUGGCUCACACCUGUAAUCCCAGUACUUUGGGAAGCCGAGGCGGGAGGAU | −28.8 |
| TPP-A549-4 | AGGCUGUAAGGCCACACGGGGUG | 18 | 374,244,03 | 374,244,95 | 374,244,63 | 374,244,85 | CCUCCAAGGCAGGCUGUAAGGCCACACGGGGUGGAAGGCCAGAGCUCCUUCUCCUCCUUCUGCUCUCUGGGUAUUGCCACCAAGCCCUGUACC | −32.8 |
| TPP-A549-5 | CCUAGGACCAUCACAGGCUGA | 16 | 888,546,38 | 888,547,12 | 888,546,82 | 888,547,02 | AACCUCCCACUGGGUCUGCUGACUGCAGGGGUGACUUGGAGUCACCUAGGACCAUCACAGGCUGAGUGAGGAUGU | −34.3 |
| TPP-A549-6 | AUACUAUCUAGCUUUGGGUAGUA | 6 | 705,117,52 | 705,118,32 | 705,117,62 | 705,117,84 | GCGUAGGAGUAACAGGCUGUAUUAUCUAGUUUAGGUGUGUAGCAGGCUAUACUAUCUAGCUUUGGGUAGUAUAUUCUAUGA | −25.3 |
| TPP-A549-7 | CAGAUGGUUGCUGAUCUGUGCA | 4 | 107,393,112 | 107,393,209 | 107,393,122 | 107,393,143 | CAGGGUGUCCCAGAUGUCUCUGCAGCUUCCUCCUUGCAUCAGAUGCCAGCAUCCAGUGACUCGAUCCAGAUGGUUGCUGAUCUGUGCAGCAGAUCCUC | −29 |
| TPP-A549-8 | AGUGUUUAAGAUCCAAGUGUUG | 4 | 911,143,10 | 911,143,99 | 911,143,68 | 911,143,89 | UGUGCUUCUAAGUGUUUAAGAUCCAAGUGUUGAUAAUGGAAGUCACAUUAUCAACAUAAUUGGUAUCUAUAGAUGCUCUUUGGAUUCCAA | −29.2 |
| TPP-A549-9 | UUGGUGGUGUACACGGAGCAG | 19 | 385,161,25 | 385,162,07 | 385,161,77 | 385,161,97 | CCGCGGAGACUUGGUGGUGUACACGGAGCAGGCGUUGUACUCGUUCAGCUGCGGCUCCAGGAACGCCACCGGCAUGGCUGCUG | −36.6 |
| TPP-A549-10 | CCAGGACUCGAACCCUCCACCC | 8 | 144,458,110 | 144,458,200 | 144,458,169 | 144,458,190 | ACAUGGGUCUGAGAGGGACUGGCACGUUGUCCAAGGCCGCACAGCUGUUAAGCAUGAAGCCAGGACUCGAACCCUCCACCCAGGCUGGAUU | −29.8 |
| 40 μM EGCG Treatment | ||||||||
| TPP-A549-11 | CUAAGACUGCAGUGAGCCUUGGU | 16 | 589,8984 | 589,9051 | 589,8994 | 589,9016 | GCCCAGGAGACUAAGACUGCAGUGAGCCUUGGUUUGCAGUGAGCCUCACUGCUUUUAGUUUCAGAGAU | −25.1 |
| TPP-A549-12 | CACCUGUAGUUCCAUCUACUCCAG | 7 | 789,447,92 | 789,44892 | 789,448,59 | 789,448,82 | UAGUGGCGUGCACCUGUAGUUCCAUCUACUCCAGAGGCUGAGGUGGGGGGAUCGUUUGAGCUUGGGAGAUGGAGGUUGCAGUGAGCAGUGAUUGUGCCACU | −38.51 |
| TPP-A549-13 | CAAGCCAUUGAUCCCAAAGUCA | 1 | 535,110,24 | 535,111,16 | 535,110,85 | 535,111,06 | CCAGAGCCCCCAAGCCAUUGAUCCCAAAGUCAUUUUAGCCCCUGUGAACGGGGCCCAGGGGCUUAGGAAGGUAGCAUGAGAAGGGAACCUGGC | −29.3 |
| TPP-A549-14 | CUCUGAACUGGGUAGAUCUGUC | 2 | 181,190,09 | 181,190,98 | 181,190,19 | 181,190,40 | AACAGGGUAGCUCUGAACUGGGUAGAUCUGUCUGUCAAGAGGAACGCUCUAGAAAGCUCAGGACUCUCCUGCACAGGGCUUAGCCCACUG | −31.6 |
| TPP-A549-9 | UUGGUGGUGUACACGGAGCAGGC | 19 | 385,161,25 | 385,162,07 | 3851,6175 | 385,161,97 | CCGCGGAGACUUGGUGGUGUACACGGAGCAGGCGUUGUACUCGUUCAGCUGCGGCUCCAGGAACGCCACCGGCAUGGCUGCUG | −36.6 |
| TPP-A549-8 | AGUGUUUAAGAUCCAAGUGUUG | 4 | 911,143,10 | 911,143,99 | 911,143,68 | 911,143,89 | UGUGCUUCUAAGUGUUUAAGAUCCAAGUGUUGAUAAUGGAAGUCACAUUAUCAACAUAAUUGGUAUCUAUAGAUGCUCUUUGGAUUCCAA | −29.2 |
| TPP-A549-7 | CAGAUGGUUGCUGAUCUGUGCA | 4 | 107,393,112 | 107,393,209 | 107,393,122 | 107,393,143 | CAGGGUGUCCCAGAUGUCUCUGCAGCUUCCUCCUUGCAUCAGAUGCCAGCAUCCAGUGACUCGAUCCAGAUGGUUGCUGAUCUGUGCAGCAGAUCCUC | −29 |
| TPP-A549-3 | CUAGGACCAUCACAGGCUGA | 16 | 888,546,38 | 888,547,12 | 888,546,83 | 888,547,02 | AACCUCCCACUGGGUCUGCUGACUGCAGGGGUGACUUGGAGUCACCUAGGACCAUCACAGGCUGAGUGAGGAUGU | −34.3 |
| TPP-A549-15 | AAUUUCUGAUCCUGGCCAUAUUC | 1 | 414,2086 | 414,2164 | 414,2096 | 414,2118 | ACAGAGGAUAGAGUUGGGCAGCAUCAGAAAGCCCAGCUUCCUCUGGAAUUUCUGAUCCUGGCCAUAUUCACUUCCUGUG | −34.5 |
| TPP-A549-16 | GCUAAAACUGGUCAAAGUGCUG | 6 | 117,578,916 | 117,579,011 | 117,578,926 | 117,578,947 | UGUUUUGGAGAAAGAAGUACAUGAUCAGCUUUUACAGCUGCACUCUAUUCAGCUGCAGCUUCAUGCUAAAACUGGUCAAAGUGCUGACUCUGGUAC | −28.5 |
| 100 μM EGCG Treatment | ||||||||
| TPP-A549-17 | AUCAGUGGUUCUCAAUGUUCUUUU | 1 | 797,2080 | 797,2153 | 797,2120 | 797,2143 | GUUAUGGGCCAGAGGUCCUGGGGACUUGAUACAUUCGAGUAUCAGUGGUUCUCAAUGUUCUUUUGGUGCACUUG | −27.5 |
| TPP-A549-11 | CUAAGACUGCAGUGAGCCUUGGU | 16 | 589,8984 | 589,9051 | 589,8994 | 589,9016 | GCCCAGGAGACUAAGACUGCAGUGAGCCUUGGUUUGCAGUGAGCCUCACUGCUUUUAGUUUCAGAGAU | −25.1 |
| TPP-A549-18 | CAUCCACCUGGUCCAGCUCGC | 16 | 317,07819 | 317,079,08 | 317,078,29 | 317,078,49 | ACCUGGGAGGGACCAAGGCUGGGAGGGUGAGAGGCCAUGCCCACAAAAGCCUCAGGCUACAUCCACCUGGUCCAGCUCGCCAAGAUGGGA | −34.2 |
| TPP-A549-19 | GCUGUUGUUGCUGUGGUUGUGGCU | 2 | 182,3300 | 182,3382 | 182,3310 | 182,3333 | UGGCUGUGAGGCUGUUGUUGCUGUGGUUGUGGCUGCAACCUUCAUUGUCAUAGAUAUAGCUGUGGCAGCUGUAGUUGUAGCCU | −34.02 |
| TPP-A549-7 | CAGAUGGUUGCUGAUCUGUGCA | 4 | 107,3931,12 | 107,393,209 | 107,393,122 | 107,393,143 | CAGGGUGUCCCAGAUGUCUCUGCAGCUUCCUCCUUGCAUCAGAUGCCAGCAUCCAGUGACUCGAUCCAGAUGGUUGCUGAUCUGUGCAGCAGAUCCUC | −29 |
| TPP-A549-20 | GCUUAGCUAAGAAUACUUAUAAUU | 3 | 997,767,29 | 997,768,24 | 997,767,39 | 997,767,62 | AGCUUGGGCUGCUUAGCUAAGAAUACUUAUAAUUGUUUCUUGAUUAUAUGCUAAACAAGGGGUGGAUUCAUGAGUUUUCUGGGAAAGGGGUAGGCA | −25.5 |
| TPP-A549-21 | GAAGCUGUUGGACUAGAAAAAAUU | 4 | 106,688,766 | 106,688,864 | 1066,888,31 | 106,688,854 | UACCCUCUGAUAGGUUUUCUCAGCUGCAGCUUCACUGUGUGCAUAACCUCACUGUGGGGCUCAAGGAAGCUGUUGGACUAGAAAAAAUUUCAUUGCUGC | −25.6 |
| TPP-A549-1 | CCAGGAUGCACGCUCGCUGGGC | 17 | 375,2068 | 375,2154 | 375,2078 | 375,2099 | CCGUUGCCACCCAGGAUGCACGCUCGCUGGGCUGGUGGGUGCCCACUCCGUUGCCACCCAGGAUGCACGCUCACUGGCCAGGGACCU | −30.7 |
| TPP-A549-22 | GUAAAGACGUUGAUGCUGCUA | 17 | 857,0265 | 857,0362 | 857,0332 | 857,0352 | AGGAGUUUCAUAUACUAUCUCUAUCAUAUAGACAGGGCAAUGUCUUCACCUCCACUUUCAGAGGAUGGUAAAGACGUUGAUGCUGCUAACUUGCUCAG | −26.22 |
| TPP-A549-26 | UCUUCAUCUGGUUUGUGAACUUUU | 17 | 569,836,22 | 569,837,13 | 569,836,32 | 569,836,55 | GCGGUUAACAUCUUCAUCUGGUUUGUGAACUUUUUACUGCUUAGGAAAUCUUAAGAUACAAAGGGCAUAUGAUCAGGGGGAUGUAUAGCCUU | −31.7 |
| Treatments Compared | No. of MicroRNAs | MicroRNAs | |
|---|---|---|---|
| Up-Regulated | Down-Regulated | ||
| Control vs. 40 μM | 115 | hsa-miR-125a-3p, hsa-miR-15b-3p, hsa-miR-548av-3p, hsa-miR-1294, hsa-miR-335-5p, hsa-miR-937-3p, hsa-miR-3118, hsa-miR-125a-5p, hsa-miR-769-3p, hsa-miR-2277-5p, hsa-miR-30c-2-3p, hsa-miR-24-1-5p, hsa-miR-181a-2-3p, hsa-miR-105-5p, hsa-miR-181c-5p, hsa-miR-365a-3p, hsa-miR-374c-5p, hsa-miR-3922-5p, hsa-miR-26b-5p, hsa-miR-449c-5p, hsa-miR-556-3p, hsa-miR-758-3p, hsa-miR-151a-5p, hsa-miR-331-5p, hsa-miR-942-5p, hsa-miR-20b-5p, hsa-miR-548a-3p, hsa-miR-103a-2-5p, hsa-miR-323b-3p, hsa-miR-421, hsa-miR-183-3p, hsa-miR-940, hsa-miR-935, hsa-miR-9-5p, hsa-let-7f-1-3p, hsa-miR-1468-5p, hsa-miR-4510, hsa-miR-5009-5p, hsa-miR-589-3p, hsa-miR-6747-3p, hsa-miR-195-5p, hsa-miR-486-3p, hsa-miR-454-3p, hsa-miR-3173-5p, hsa-miR-543, hsa-miR-125b-2-3p, hsa-miR-155-5p, hsa-miR-3179, hsa-miR-1976, hsa-miR-598-3p, hsa-miR-548al, hsa-miR-6853-3p, hsa-miR-6733-5p | hsa-miR-548o-3p, hsa-miR-96-5p, hsa-miR-185-3p, hsa-miR-1299 hsa-miR-22-5p, hsa-miR-195-3p, hsa-miR-653-5p, hsa-miR-425-3p, hsa-miR-215-5p, hsa-miR-7-1-3p, hsa-miR-132-5p, hsa-miR-424-5p, hsa-miR-3613-5p, hsa-miR-30b-5p, hsa-miR-548f-5p, hsa-miR-26a-2-3p, hsa-miR-18a-3p, hsa-miR-30b-3p, hsa-miR-654-3p, hsa-miR-140-5p, hsa-miR-6720-3p, hsa-miR-493-5p, hsa-miR-122-5p, hsa-miR-548n, hsa-miR-580-3p, hsa-miR-6723-5p, hsa-miR-1303, hsa-miR-597-3p, hsa-miR-1293, hsa-miR-10a-3p, hsa-miR-4473, hsa-miR-3163, hsa-miR-3191-5p, hsa-miR-365a-5p, hsa-miR-100-3p, hsa-miR-191-5p, hsa-miR-130b-3p, hsa-miR-20a-3p, hsa-miR-3134, hsa-miR-23a-5p, hsa-miR-556-5p, hsa-miR-4488, hsa-miR-491-5p, hsa-miR-378i, hsa-miR-3942-5p, hsa-miR-548j-3p, hsa-miR-6806-3p, hsa-miR-200a-3p, hsa-miR-212-3p, hsa-miR-105-3p, hsa-miR-1273g-3p, hsa-miR-34a-3p, hsa-miR-342-3p, hsa-miR-192-3p, hsa-miR-29c-5p, hsa-let-7d-5p, hsa-miR-1305, hsa-miR-146b-3p, hsa-miR-578, hsa-miR-6811-5p, hsa-miR-552-3p, hsa-miR-4755-3p, hsa-miR-3127-5p |
| 40 Control vs. 100 μM | 121 | hsa-miR-548ah-3p, hsa-miR-96-5p, hsa-miR-500a-3p, hsa-miR-185-3p, hsa-miR-22-5p, hsa-miR-7706, hsa-miR-30e-3p, hsa-miR-532-5p, hsa-miR-502-3p, hsa-miR-1299, hsa-miR-4677-3p, hsa-miR-582-5p, hsa-miR-195-3p, hsa-miR-424-5p, hsa-miR-425-3p, hsa-miR-3613-5p, hsa-miR-653-5p, hsa-miR-424-3p, hsa-miR-152-5p, hsa-miR-32-3p, hsa-miR-18a-3p, hsa-miR-548n, hsa-miR-493-5p, hsa-miR-30b-3p, hsa-miR-618, hsa-miR-654-3p, hsa-miR-548f-5p, hsa-miR-140-5p, hsa-miR-92a-1-5p, hsa-miR-146b-3p, hsa-miR-548g-3p, hsa-miR-26a-2-3p, hsa-miR-10a-3p, hsa-miR-598-3p, hsa-miR-4502, hsa-miR-423-3p, hsa-miR-191-5p, hsa-miR-6720-3p, hsa-miR-3664-3p, hsa-miR-130b-5p, hsa-miR-3150b-3p, hsa-miR-4473, hsa-miR-2682-3p, hsa-miR-365a-5p, hsa-miR-215-5p, hsa-miR-4742-3p, hsa-miR-6511a-3p, hsa-miR-28-5p, hsa-miR-580-3p, hsa-miR-3942-5p, hsa-miR-6775-3p, hsa-miR-6818-3p, hsa-miR-3591-5p, hsa-miR-625-3p, hsa-miR-6868-3p, hsa-miR-597-3p, hsa-miR-381-3p, hsa-miR-127-3p, hsa-miR-1229-3p, hsa-miR-200a-3p, hsa-miR-23a-5p, hsa-miR-3120-3p, hsa-miR-433-3p, hsa-miR-4677-5p, hsa-miR-655-3p, hsa-miR-5091, hsa-miR-627-3p, hsa-miR-548aj-5p | hsa-miR-3184-5p, hsa-miR-548av-3p, hsa-miR-27a-3p, hsa-let-7d-3p, hsa-miR-548f-3p, hsa-miR-361-5p, hsa-miR-365a-3p, hsa-miR-3607-3p, hsa-miR-584-5p, hsa-miR-3922-5p, hsa-miR-148b-5p, hsa-miR-7641, hsa-miR-181b-3p, hsa-miR-3158-3p, hsa-miR-105-5p, hsa-miR-4796-5p, hsa-miR-1-3p, hsa-miR-151a-5p, hsa-miR-374a-5p, hsa-miR-1293, hsa-miR-4775, hsa-miR-574-3p, hsa-miR-331-5p, hsa-miR-107, hsa-miR-4717-5p, hsa-miR-363-3p, hsa-let-7f-2-3p, hsa-miR-454-5p, hsa-miR-103a-2-5p, hsa-miR-374c-5p, hsa-miR-6783-3p, hsa-miR-1185-2-3p, hsa-miR-935, hsa-miR-1468-5p, hsa-miR-744-3p, hsa-miR-3131, hsa-miR-4762-3p, hsa-miR-873-5p, hsa-miR-3923, hsa-let-7a-2-3p, hsa-miR-769-3p, hsa-miR-1249-5p, hsa-miR-142-5p, hsa-miR-181c-5p, hsa-miR-3179, hsa-miR-33a-5p, hsa-miR-3688-3p, hsa-miR-3944-3p, hsa-miR-410-3p, hsa-miR-4684-5p, hsa-miR-548ay-3p, hsa-miR-636, hsa-miR-653-3p, hsa-miR-6843-3p |
| Control vs. 100 μM | 134 | hsa-miR-125a-3p, hsa-miR-500a-3p, hsa-miR-7706, hsa-miR-15b-3p, hsa-miR-598-3p, hsa-miR-532-5p, hsa-miR-502-3p, hsa-miR-30e-3p, hsa-miR-181a-2-3p, hsa-miR-1294, hsa-miR-24-1-5p, hsa-miR-424-3p, hsa-miR-335-5p, hsa-miR-32-3p, hsa-miR-125a-5p, hsa-miR-618, hsa-miR-3664-3p, hsa-miR-3118, hsa-miR-582-5p, hsa-miR-4677-3p, hsa-miR-2277-5p, hsa-miR-30c-2-3p, hsa-miR-152-5p, hsa-miR-92a-1-5p, hsa-miR-421, hsa-miR-937-3p, hsa-miR-423-3p, hsa-miR-26b-5p, hsa-miR-548g-3p, hsa-miR-130b-5p, hsa-miR-625-3p, hsa-miR-9-5p, hsa-miR-556-3p, hsa-miR-758-3p, hsa-miR-769-3p, hsa-miR-5009-5p hsa-miR-2682-3p, hsa-miR-454-3p, hsa-miR-4502, hsa-miR-222-3p, hsa-miR-26a-1-3p, hsa-miR-3140-3p, hsa-miR-3591-5p, hsa-miR-548al, hsa-miR-940, hsa-miR-1255a, hsa-let-7f-1-3p, hsa-miR-146b-3p, hsa-miR-6868-3p, hsa-miR-548x-3p, hsa-miR-1285-5p, hsa-miR-1304-3p, hsa-miR-2116-3p, hsa-miR-28-5p, hsa-miR-543, hsa-miR-1229-3p, hsa-miR-1290, hsa-miR-155-5p, hsa-miR-181b-2-3p, hsa-miR-1976 hsa-miR-3120-3p, hsa-miR-3129-5p, hsa-miR-3174, hsa-miR-323b-3p, hsa-miR-432-5p, hsa-miR-548a-3p, hsa-miR-589-3p, hsa-miR-655-3p hsa-miR-6747-3p | hsa-miR-548o-3p, hsa-miR-3184-5p, hsa-miR-27a-3p, hsa-miR-548f-3p, hsa-let-7d-3p, hsa-miR-1293, hsa-miR-7641, hsa-miR-3607-3p, hsa-miR-132-5p, hsa-miR-7-1-3p, hsa-miR-3158-3p, hsa-miR-361-5p, hsa-miR-6723-5p, hsa-miR-1-3p, hsa-miR-148b-5p, hsa-miR-30b-5p, hsa-miR-584-5p, hsa-miR-491-5p, hsa-miR-181b-3p, hsa-miR-574-3p, hsa-miR-1303, hsa-let-7a-2-3p, hsa-miR-215-5p, hsa-miR-548q, hsa-miR-629-3p, hsa-miR-1262, hsa-miR-107, hsa-miR-374a-5p, hsa-let-7d-5p, hsa-miR-3688-3p, hsa-miR-4793-3p, hsa-miR-744-3p, hsa-miR-4796-5p, hsa-miR-20a-3p, hsa-miR-122-5p, hsa-miR-6761-5p, hsa-miR-6806-3p, hsa-miR-135b-5p, hsa-miR-4775, hsa-miR-450a-1-3p, hsa-miR-142-5p, hsa-miR-378i, hsa-miR-7705, hsa-miR-548ay-3p, hsa-miR-548j-3p, hsa-miR-1273g-3p, hsa-miR-130b-3p, hsa-miR-4488, hsa-miR-345-5p, hsa-let-7f-2-3p, hsa-miR-192-3p, hsa-miR-1249-5p, hsa-miR-136-5p, hsa-miR-184, hsa-miR-3127-5p, hsa-miR-3163, hsa-miR-3191-5p, hsa-miR-3654,hsa-miR-3691-5p, hsa-miR-3944-3p, hsa-miR-4477b, hsa-miR-4787-3p, hsa-miR-485-5p, hsa-miR-578, hsa-miR-6737-3p, hsa-miR-6843-3p |
| Treatment Compared | No. of MicroRNAs | Putative Novel Micro RNA ID | Log2 Fold Change | microRNA Sequences | Chromosome Number | Precursor Sequence Start | Precursor Sequence End | Mature Sequence Start | Mature Sequence End | Precursor Sequence | MFE (Kcal/mol) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Up-Regulated | Down-Regulated | |||||||||||
| Control vs. 40 μM | 4 | TPP-A549-26 | 2.52 | CUCUGAACUGGGUAGAUCUGUC | 2 | 181,190,09 | 181,190,98 | 181,190,19 | 181,190,40 | AACAGGGTAGCTCTGAACTGGGTAGATCTGTCTGTCAAGAGGAACGCTCTAGAAAGCTCAGGACTCTCCTGCACAGGGCTTAGCCCACTG | −31.6 | |
| TPP-A549-24 | -2.5 | AAUAGGAAUUGAGAGAAGUG | 4 | 107,162,755 | 107,162,845 | 107,162,765 | 107,162,784 | CTCATGTTTTACTTCCTTTTCCTATTTTGTTACACTAGCTAGGGCTTCTAGTAGAGTAATGAATAGGAATTGAGAGAAGTGATATTTTGGC | −27.5 | |||
| TPP-A549-27 | -3.34 | CUUAGAAUUCCGAUGCUGGGG | 22 | 267,618,39 | 267,619,28 | 267,618,98 | 267,619,18 | AGATTCTGGACTTAGAATTCCGATGCTGGGGCCAGGCACAGTGGCTCACACCTGTAATCCCAGTACTTTGGGAAGCCGAGGCGGGAGGAT | −28.8 | |||
| TPP-A549-28 | -2.23 | GAAAAACCUGAGCUGAUGGAC | 5 | 140,056,955 | 140,057,037 | 140,057,007 | 140,057,027 | TCTAGATGATGAAAAACCTGAGCTGATGGACAATGCCCGTAAGTGTATTGTGATATTGCAGCTCAGGTTTTTCATTAATGTAA | −36.3 | |||
| 40 vs. 100 μM | 4 | TPP-A549-29 | 2.2 | GAGGUAGAGCAAGGGCCCCU | 13 | 394,145,14 | 394,146,06 | 394,145,24 | 394,145,43 | GGCTGGAGTTGAGGTAGAGCAAGGGCCCCTGCATTTGGTGTTATCAGTCTTCTGGGACTTTCTTGGGGTACTGTTTTCACTTCTTCTTCTTAT | −27.2 | |
| TPP-A549-30 | 2.6 | UUCAGGUUGAAGCUCUUGCACA | 14 | 102,442,311 | 102,442,392 | 102,442,321 | 102,442,342 | GGGATGAGTCTTCAGGTTGAAGCTCTTGCACAGCTGGCTCTCTCCTAGCTGTGTAAGAACCTCTGGCCTGGGTGCACAGCCA | −38 | |||
| TPP-A549-31 | −2.32 | CCAGCCUACUGGAGGAUAAGAGGA | 8 | 102,442,311 | 102,4423,92 | 102,442,321 | 102,442,342 | GGGATGAGTCTTCAGGTTGAAGCTCTTGCACAGCTGGCTCTCTCCTAGCTGTGTAAGAACCTCTGGCCTGGGTGCACAGCCA | −38 | |||
| TPP-A549-32 | −2.22 | CUAGGACCAUCACAGGCUGA | 16 | 983,936,55 | 983,937,34 | 983,937,01 | 983,937,24 | CTCTGGTTCCCCAGCCTACTGGAGGATAAGAGGATATAAAGGTCTCTTATCCTCCAGTAGACTAGGGAGCCAGAGCTGGT | −53.7 | |||
| Control vs. 100 μM | 3 | TPP-A549-33 | 2.43 | GAGGUAGAGCAAGGGCCCCU | 13 | 394,145,14 | 394,146,06 | 394,145,24 | 394,145,43 | GGCTGGAGTTGAGGTAGAGCAAGGGCCCCTGCATTTGGTGTTATCAGTCTTCTGGGACTTTCTTGGGGTACTGTTTTCACTTCTTCTTCTTAT | −27.2 | |
| TPP-A549-23 | −4.2 | CUUGCAAUUACUUUUGCACCAACU | 4 | 158,492,16 | 158,493,08 | 158,492,26 | 158,492,49 | TAAATTATTAGGTTGGTTCAAAAATAATTGTGGTTTTGCCATTCCTTTCGGTGGCAAAACTTGCAATTACTTTTGCACCAACTTAAATATATA | −37.4 | |||
| TPP-A549-34 | −4.2 | UAGGUGGAUUUGUGUGAUGAGCU | 9 | 195,7951 | 195,8044 | 195,7961 | 195,7983 | AGGCCTGCCATAGGTGGATTTGTGTGATGAGCTAAGGAGTTTCCCAAGCAGATATCATCCTAATGGAAAAGAGAACCAAGCACAAGCTCGGTCA | −25.5 | |||
| MicroRNAs | KEGG Pathways Common in TargetScan and miRDB | PANTHER Pathways Common in TargetScan and miRDB | KEGG Pathway Common in all the MicroRNAs | PANTHER Pathways Common in all the microRNAs |
|---|---|---|---|---|
| Hsa-miR-21-5p | MAPK signaling pathway, Regulation of actin cytoskeleton, Chemokine signaling pathway, Pathways in cancer, Neurotrophin signaling pathway | P00021: FGF signaling pathway, P00005: Angiogenesis, P04393: Ras Pathway | MAPK signaling pathway | Nil |
| Hsa-miR-548o-5p | Small cell lung cancer, Colorectal cancer, Pancreatic cancer, Renal cell carcinoma, T cell receptor signaling pathway, the Hedgehog signaling pathway, Long-term potentiation, TGF-beta signaling pathway, ErbB signaling pathway, Endometrial cancer, B cell receptor signaling pathway, Acute myeloid leukemia, Glioma, Non-small cell lung cancer, MAPK signaling pathway, Axon guidance, Apoptosis, Wnt signaling pathway | P00057: Wnt signaling pathway | ||
| Hsa-miR-181c | Neurotrophin signaling pathway, Axon guidance, Long-term potentiation, Dorso-ventral axis formation, TGF-beta signaling pathway, Pathways in cancer, MAPK signaling, pathway Colorectal cancer, Chronic myeloid leukemia, T cell receptor signaling pathway, Renal cell carcinoma, Glioma, ErbB signaling pathway, Adherens junction, Prostate cancer, Acute myeloid leukemia, Pancreatic cancer, Endometrial cancer, Phosphatidylinositol signaling system | P00021: FGF signaling pathway, P00018: EGF receptor signaling pathway, P00057: Wnt signaling pathway P00032: Insulin/IGF pathway-mitogen activated protein kinase/MAP kinase cascade, P00059: p53 pathway | ||
| Hsa-miR-212-5p | Colorectal cancer, Regulation of actin cytoskeleton, MAPK signaling pathway, Long-term potentiation | Nil |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhardwaj, V.; Mandal, A.K.A. Next-Generation Sequencing Reveals the Role of Epigallocatechin-3-Gallate in Regulating Putative Novel and Known microRNAs Which Target the MAPK Pathway in Non-Small-Cell Lung Cancer A549 Cells. Molecules 2019, 24, 368. https://doi.org/10.3390/molecules24020368
Bhardwaj V, Mandal AKA. Next-Generation Sequencing Reveals the Role of Epigallocatechin-3-Gallate in Regulating Putative Novel and Known microRNAs Which Target the MAPK Pathway in Non-Small-Cell Lung Cancer A549 Cells. Molecules. 2019; 24(2):368. https://doi.org/10.3390/molecules24020368
Chicago/Turabian StyleBhardwaj, Vaishali, and Abul Kalam Azad Mandal. 2019. "Next-Generation Sequencing Reveals the Role of Epigallocatechin-3-Gallate in Regulating Putative Novel and Known microRNAs Which Target the MAPK Pathway in Non-Small-Cell Lung Cancer A549 Cells" Molecules 24, no. 2: 368. https://doi.org/10.3390/molecules24020368