Promutagenicity of 8-Chloroguanine, A Major Inflammation-Induced Halogenated DNA Lesion
Abstract
1. Introduction
2. Results and Discussion
2.1. Steady-State Kinetic Studies
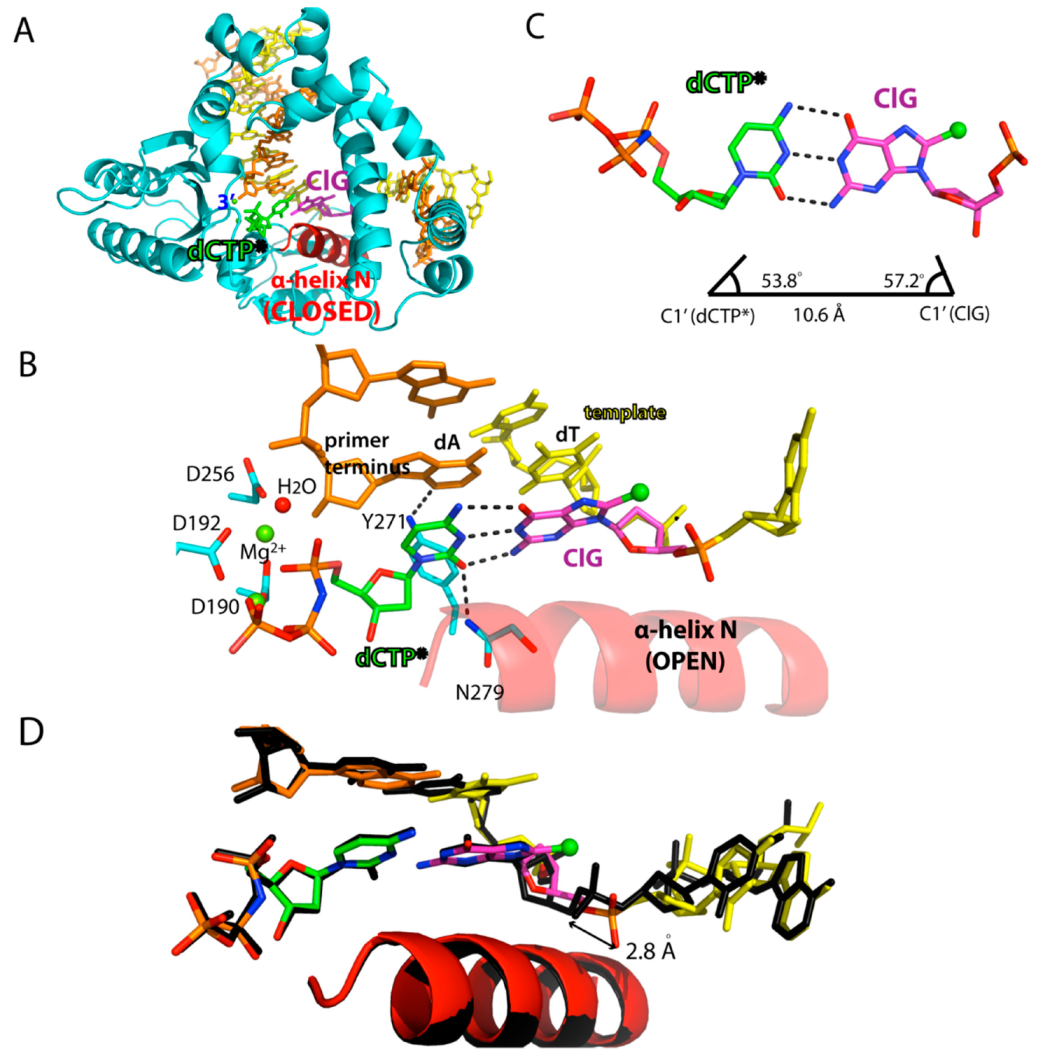
2.2. Structure of A Single-Nucleotide Gapped Binary Complex of Polβ with Templating ClG
2.3. Ternary Structure of Polβ with Templating ClG Paired with an Incoming dCTP Analog
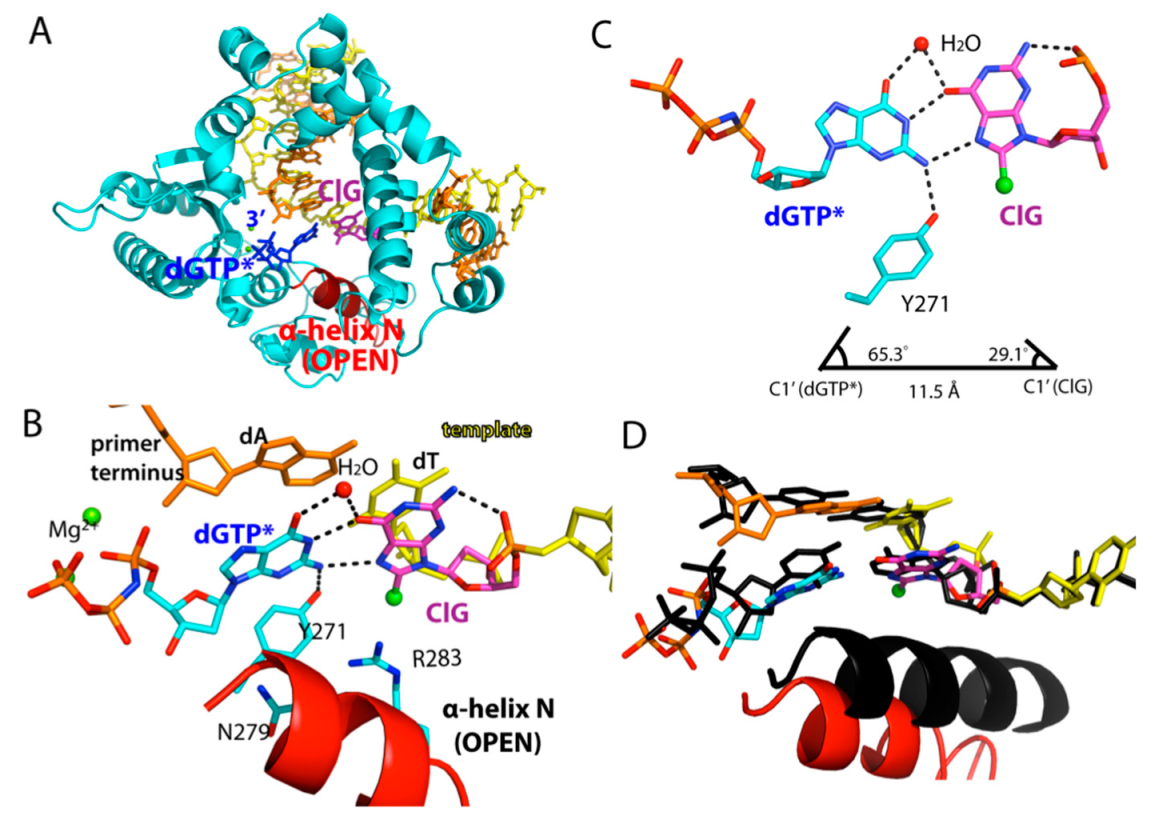
2.4. Ternary Structure of Polβ with Templating ClG Paired with an Incoming dGTP Analog
3. Materials and Methods
3.1. Synthesis of ClG (8-chloro-dG) Phosphoramidite
3.2. DNA Sequences
3.3. Steady-State Kinetics
3.4. Protein Expression and Purification, Protein-DNA Co-crystallization
3.5. Data Collection and Refinement
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jena, N.R. DNA damage by reactive species: Mechanisms, mutation and repair. J. Biosci 2012, 37, 503–517. [Google Scholar] [CrossRef] [PubMed]
- Krasner, S.W. The formation and control of emerging disinfection by-products of health concern. Philos. Trans. A Math. Phys. Eng. Sci 2009, 367, 4077–4095. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.J.; Li, W.W.; Jiang, H.; Yu, H.Q. Fates of Chemical Elements in Biomass during Its Pyrolysis. Chem. Rev. 2017, 117, 6367–6398. [Google Scholar] [CrossRef] [PubMed]
- Rivera-Utrilla, J.; Sanchez-Polo, M.; Ferro-Garcia, M.A.; Prados-Joya, G.; Ocampo-Perez, R. Pharmaceuticals as emerging contaminants and their removal from water. A review. Chemosphere 2013, 93, 1268–1287. [Google Scholar] [CrossRef] [PubMed]
- Kogevinas, M.; Villanueva, C.M.; Font-Ribera, L.; Liviac, D.; Bustamante, M.; Espinoza, F.; Nieuwenhuijsen, M.J.; Espinosa, A.; Fernandez, P.; DeMarini, D.M.; et al. Genotoxic effects in swimmers exposed to disinfection by-products in indoor swimming pools. Environ. Health Perspect. 2010, 118, 1531–1537. [Google Scholar] [CrossRef] [PubMed]
- Richardson, S.D.; DeMarini, D.M.; Kogevinas, M.; Fernandez, P.; Marco, E.; Lourencetti, C.; Balleste, C.; Heederik, D.; Meliefste, K.; McKague, A.B.; et al. What’s in the pool? A comprehensive identification of disinfection by-products and assessment of mutagenicity of chlorinated and brominated swimming pool water. Environ. Health Perspect. 2010, 118, 1523–1530. [Google Scholar] [CrossRef] [PubMed]
- Richardson, S.D.; Plewa, M.J.; Wagner, E.D.; Schoeny, R.; Demarini, D.M. Occurrence, genotoxicity, and carcinogenicity of regulated and emerging disinfection by-products in drinking water: A review and roadmap for research. Mutat. Res. 2007, 636, 178–242. [Google Scholar] [CrossRef]
- Sasada, T.; Hinoi, T.; Saito, Y.; Adachi, T.; Takakura, Y.; Kawaguchi, Y.; Sotomaru, Y.; Sentani, K.; Oue, N.; Yasui, W.; et al. Chlorinated Water Modulates the Development of Colorectal Tumors with Chromosomal Instability and Gut Microbiota in Apc-Deficient Mice. PLoS ONE 2015, 10, e0132435. [Google Scholar] [CrossRef]
- McKenna, S.M.; Davies, K.J. The inhibition of bacterial growth by hypochlorous acid. Possible role in the bactericidal activity of phagocytes. Biochem. J. 1988, 254, 685–692. [Google Scholar] [CrossRef]
- Pattison, D.I.; Davies, M.J.; Hawkins, C.L. Reactions and reactivity of myeloperoxidase-derived oxidants: Differential biological effects of hypochlorous and hypothiocyanous acids. Free Radic. Res. 2012, 46, 975–995. [Google Scholar] [CrossRef]
- Alfakry, H.; Malle, E.; Koyani, C.N.; Pussinen, P.J.; Sorsa, T. Neutrophil proteolytic activation cascades: A possible mechanistic link between chronic periodontitis and coronary heart disease. Innate Immun. 2016, 22, 85–99. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Cui, Y.; Niedernhofer, L.J.; Wang, Y. Occurrence, Biological Consequences, and Human Health Relevance of Oxidative Stress-Induced DNA Damage. Chem. Res. Toxicol. 2016, 29, 2008–2039. [Google Scholar] [CrossRef] [PubMed]
- Davies, M.J. Myeloperoxidase-derived oxidation: Mechanisms of biological damage and its prevention. J. Clin. Biochem. Nutr. 2011, 48, 8–19. [Google Scholar] [CrossRef] [PubMed]
- Kato, Y. Neutrophil myeloperoxidase and its substrates: Formation of specific markers and reactive compounds during inflammation. J. Clin. Biochem. Nutr. 2016, 58, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Henderson, J.P.; Byun, J.; Williams, M.V.; Mueller, D.M.; McCormick, M.L.; Heinecke, J.W. Production of brominating intermediates by myeloperoxidase. A transhalogenation pathway for generating mutagenic nucleobases during inflammation. J. Biol. Chem. 2001, 276, 7867–7875. [Google Scholar] [CrossRef] [PubMed]
- Masuda, M.; Suzuki, T.; Friesen, M.D.; Ravanat, J.L.; Cadet, J.; Pignatelli, B.; Nishino, H.; Ohshima, H. Chlorination of guanosine and other nucleosides by hypochlorous acid and myeloperoxidase of activated human neutrophils. Catalysis by nicotine and trimethylamine. J. Biol. Chem. 2001, 276, 40486–40496. [Google Scholar] [CrossRef]
- Badouard, C.; Masuda, M.; Nishino, H.; Cadet, J.; Favier, A.; Ravanat, J.L. Detection of chlorinated DNA and RNA nucleosides by HPLC coupled to tandem mass spectrometry as potential biomarkers of inflammation. J. Chromatogr B. Anal. Technol. Biomed. Life Sci. 2005, 827, 26–31. [Google Scholar] [CrossRef]
- Suzuki, T.; Friesen, M.D.; Ohshima, H. Identification of products formed by reaction of 3′,5′-di-O-acetyl-2′-deoxyguanosine with hypochlorous acid or a myeloperoxidase-H2O2-Cl-system. Chem. Res. Toxicol 2003, 16, 382–389. [Google Scholar] [CrossRef]
- Nauseef, W.M. Myeloperoxidase in human neutrophil host defence. Cell Microbiol 2014, 16, 1146–1155. [Google Scholar] [CrossRef]
- Winterbourn, C.C.; Kettle, A.J. Redox reactions and microbial killing in the neutrophil phagosome. Antioxid. Redox Signal. 2013, 18, 642–660. [Google Scholar] [CrossRef]
- Bouvard, V.; Baan, R.; Straif, K.; Grosse, Y.; Secretan, B.; El Ghissassi, F.; Benbrahim-Tallaa, L.; Guha, N.; Freeman, C.; Galichet, L.; et al. A review of human carcinogens—Part B: Biological agents. Lancet Oncol. 2009, 10, 321–322. [Google Scholar] [CrossRef]
- Whitcomb, D.C. Inflammation and Cancer V. Chronic pancreatitis and pancreatic cancer. Am. J. Physiol. Gastrointest Liver Physiol. 2004, 287, G315–G319. [Google Scholar] [CrossRef]
- Ohshima, H.; Tatemichi, M.; Sawa, T. Chemical basis of inflammation-induced carcinogenesis. Arch. Biochem. Biophys. 2003, 417, 3–11. [Google Scholar] [CrossRef]
- Ohnishi, S.; Murata, M.; Kawanishi, S. DNA damage induced by hypochlorite and hypobromite with reference to inflammation-associated carcinogenesis. Cancer Lett. 2002, 178, 37–42. [Google Scholar] [CrossRef]
- Gungor, N.; Knaapen, A.M.; Munnia, A.; Peluso, M.; Haenen, G.R.; Chiu, R.K.; Godschalk, R.W.; van Schooten, F.J. Genotoxic effects of neutrophils and hypochlorous acid. Mutagenesis 2010, 25, 149–154. [Google Scholar] [CrossRef]
- Sassa, A.; Kamoshita, N.; Matsuda, T.; Ishii, Y.; Kuraoka, I.; Nohmi, T.; Ohta, T.; Honma, M.; Yasui, M. Miscoding properties of 8-chloro-2′-deoxyguanosine, a hypochlorous acid-induced DNA adduct, catalysed by human DNA polymerases. Mutagenesis 2013, 28, 81–88. [Google Scholar] [CrossRef]
- Kronberg, L.; Asplund, D.; Maki, J.; Sjoholm, R. Reaction of mucochloric and mucobromic acids with adenosine and cytidine: Formation of chloro- and bromopropenal derivatives. Chem. Res. Toxicol. 1996, 9, 1257–1263. [Google Scholar] [CrossRef]
- Valinluck, V.; Wu, W.; Liu, P.; Neidigh, J.W.; Sowers, L.C. Impact of cytosine 5-halogens on the interaction of DNA with restriction endonucleases and methyltransferase. Chem. Res. Toxicol. 2006, 19, 556–562. [Google Scholar] [CrossRef]
- Theruvathu, J.A.; Kim, C.H.; Darwanto, A.; Neidigh, J.W.; Sowers, L.C. pH-Dependent configurations of a 5-chlorouracil-guanine base pair. Biochemistry 2009, 48, 11312–11318. [Google Scholar] [CrossRef][Green Version]
- Lao, V.V.; Herring, J.L.; Kim, C.H.; Darwanto, A.; Soto, U.; Sowers, L.C. Incorporation of 5-chlorocytosine into mammalian DNA results in heritable gene silencing and altered cytosine methylation patterns. Carcinogenesis 2009, 30, 886–893. [Google Scholar] [CrossRef]
- Kim, C.H.; Darwanto, A.; Theruvathu, J.A.; Herring, J.L.; Sowers, L.C. Polymerase incorporation and miscoding properties of 5-chlorouracil. Chem. Res. Toxicol 2010, 23, 740–748. [Google Scholar] [CrossRef][Green Version]
- Steenken, S.S. How easily oxidizable is DNA? One-electron reduction potentials of adenosine and guanosine radicals in aqueous solution. J. Am. Chem. Soc. 1997, 119, 617–618. [Google Scholar] [CrossRef]
- Kanvah, S.; Schuster, G.B. The sacrificial role of easily oxidizable sites in the protection of DNA from damage. Nucleic Acids Res. 2005, 33, 5133–5138. [Google Scholar] [CrossRef]
- Hamm, M.L.; Rajguru, S.; Downs, A.M.; Cholera, R. Base pair stability of 8-chloro- and 8-iodo-2′-deoxyguanosine opposite 2′-deoxycytidine: Implications regarding the bioactivity of 8-oxo-2′-deoxyguanosine. J. Am. Chem. Soc. 2005, 127, 12220–12221. [Google Scholar] [CrossRef]
- Asahi, T.; Nakamura, Y.; Kato, Y.; Osawa, T. Specific role of taurine in the 8-brominated-2′-deoxyguanosine formation. Arch. Biochem. Biophys. 2015, 586, 45–50. [Google Scholar] [CrossRef]
- Esposito, V.; Randazzo, A.; Piccialli, G.; Petraccone, L.; Giancola, C.; Mayol, L. Effects of an 8-bromodeoxyguanosine incorporation on the parallel quadruplex structure [d(TGGGT)]4. Org. Biomol. Chem. 2004, 2, 313–318. [Google Scholar] [CrossRef]
- Beard, W.A.; Wilson, S.H. Structure and mechanism of DNA polymerase Beta. Chem. Rev. 2006, 106, 361–382. [Google Scholar] [CrossRef]
- Koag, M.C.; Kou, Y.; Ouzon-Shubeita, H.; Lee, S. Transition-state destabilization reveals how human DNA polymerase beta proceeds across the chemically unstable lesion N7-methylguanine. Nucleic Acids Res. 2014, 42, 8755–8766. [Google Scholar] [CrossRef][Green Version]
- Kou, Y.; Koag, M.C.; Lee, S. N7 methylation alters hydrogen-bonding patterns of guanine in duplex DNA. J. Am. Chem. Soc. 2015, 137, 14067–14070. [Google Scholar] [CrossRef]
- Freudenthal, B.D.; Beard, W.A.; Wilson, S.H. DNA polymerase minor groove interactions modulate mutagenic bypass of a templating 8-oxoguanine lesion. Nucleic Acids Res. 2013, 41, 1848–1858. [Google Scholar] [CrossRef]
- Hsu, G.W.; Ober, M.; Carell, T.; Beese, L.S. Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase. Nature 2004, 431, 217–221. [Google Scholar] [CrossRef]
- Koag, M.C.; Min, K.; Lee, S. Structural basis for promutagenicity of 8-halogenated guanine. J. Biol. Chem. 2014, 289, 6289–6298. [Google Scholar] [CrossRef]
- Fujikawa, K.; Yakushiji, H.; Nakabeppu, Y.; Suzuki, T.; Masuda, M.; Ohshima, H.; Kasai, H. 8-Chloro-dGTP, a hypochlorous acid-modified nucleotide, is hydrolyzed by hMTH1, the human MutT homolog. FEBS Lett. 2002, 512, 149–151. [Google Scholar] [CrossRef]
- Koag, M.C.; Lee, S. Metal-dependent conformational activation explains highly promutagenic replication across O6-methylguanine by human DNA polymerase beta. J. Am. Chem. Soc. 2014, 136, 5709–5721. [Google Scholar] [CrossRef]
- Batra, V.K.; Shock, D.D.; Beard, W.A.; McKenna, C.E.; Wilson, S.H. Binary complex crystal structure of DNA polymerase beta reveals multiple conformations of the templating 8-oxoguanine lesion. Proc. Natl. Acad. Sci. USA 2012, 109, 113–118. [Google Scholar] [CrossRef]
- Batra, V.K.; Beard, W.A.; Shock, D.D.; Pedersen, L.C.; Wilson, S.H. Structures of DNA polymerase beta with active-site mismatches suggest a transient abasic site intermediate during misincorporation. Mol. Cell 2008, 30, 315–324. [Google Scholar] [CrossRef]
- Batra, V.K.; Beard, W.A.; Pedersen, L.C.; Wilson, S.H. Structures of DNA Polymerase Mispaired DNA Termini Transitioning to Pre-catalytic Complexes Support an Induced-Fit Fidelity Mechanism. Structure 2016, 24, 1863–1875. [Google Scholar] [CrossRef]
- Batra, V.K.; Beard, W.A.; Shock, D.D.; Krahn, J.M.; Pedersen, L.C.; Wilson, S.H. Magnesium-induced assembly of a complete DNA polymerase catalytic complex. Structure 2006, 14, 757–766. [Google Scholar] [CrossRef]
- Freudenthal, B.D.; Beard, W.A.; Shock, D.D.; Wilson, S.H. Observing a DNA polymerase choose right from wrong. Cell 2013, 154, 157–168. [Google Scholar] [CrossRef]
- Sawaya, M.R.; Prasad, R.; Wilson, S.H.; Kraut, J.; Pelletier, H. Crystal structures of human DNA polymerase beta complexed with gapped and nicked DNA: Evidence for an induced fit mechanism. Biochemistry 1997, 36, 11205–11215. [Google Scholar] [CrossRef]
- Bertram, J.G.; Oertell, K.; Petruska, J.; Goodman, M.F. DNA polymerase fidelity: Comparing direct competition of right and wrong dNTP substrates with steady state and pre-steady state kinetics. Biochemistry 2010, 49, 20–28. [Google Scholar] [CrossRef]
- Otwinowski, Z.; Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzym. 1997, 276, 307–326. [Google Scholar]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta. Cryst. D. Biol. Cryst. 2004, 60, 2126–2132. [Google Scholar] [CrossRef]
- Adams, P.D.; Afonine, P.V.; Bunkoczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta. Cryst. D. Biol. Cryst. 2010, 66, 213–221. [Google Scholar] [CrossRef]
- Davis, I.W.; Leaver-Fay, A.; Chen, V.B.; Block, J.N.; Kapral, G.J.; Wang, X.; Murray, L.W.; Arendall, W.B., 3rd; Snoeyink, J.; Richardson, J.S.; et al. MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007, 35, W375–W383. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |


| Template:dNTP (Metal Ion) | Km (μM) | kcat (10−3 s−1) | kcat/Km (10−3 s−1·μM−1) | fa |
|---|---|---|---|---|
| dG:dCTP (Mg2+) | 0.59 ± 0.03 | 20.38 ± 0.50 | 34.54 | 1 |
| ClG:dCTP (Mg2+) | 6.52 ± 0.81 | 3.59 ± 0.23 | 0.55 | 1.6 × 10−2 |
| ClG:dGTP (Mg2+) | 22.96 ± 0.23 | 0.87 ± 0.05 | 0.038 | 1.1 × 10−3 |
| ClG:dCTP (Mn2+) | 1.31 ± 0.22 | 16.52 ± 1.01 | 12.61 | 3.7 × 10−1 |
| ClG:dGTP (Mn2+) | 3.92 ± 0.09 | 4.52 ± 0.34 | 1.15 | 3.3 × 10−2 |
| PDB Code | ClG-Gapped Binary 6CLY | ClG:C-Mg2+ Ternary 6CPQ | ClG:G-Mg2+ Ternary 6CRH |
|---|---|---|---|
| Space Group | P21 | P21 | P21 |
| Cell constants | |||
| a (Å) b c α (°) β γ | 54.772 79.668 54.967 90.00 105.60 90.00 | 50.829 80.243 55.836 90.00 107.80 90.00 | 54.985 79.622 55.179 90.00 107.88 90.00 |
| Resolution (Å) a | 20–2.18 (2.22–2.18) | 20–1.93 (1.96–1.93) | 20–2.33 (2.37–2.33) |
| Rmerge b (%) | 0.090 (0.548) | 0.051 (0.247) | 0.096 (0.488) |
| <I/σ> | 20.8 (1.86) | 31.2 (4.31) | 20.1 (2.36) |
| Completeness (%) | 100 (99.9) | 100 (100) | 100 (100) |
| Redundancy | 5.4 (5.1) | 4.7 (4.2) | 5.6 (5.4) |
| Refinement | P21 | P21 | P21 |
| Rwork c/Rfree d (%) | 21.8/ 26.8 | 19.1/ 23.9 | 19.7/ 24.3 |
| Unique reflections | 23552 | 32119 | 19481 |
| mean B factor (Å2) | |||
| Protein | 36.3 | 23.3 | 41.2 |
| Ligand | 34.5 | 27.7 | 37.6 |
| Solvent | 33.3 | 32.6 | 38.0 |
| Ramachandran plot | |||
| Most favored (%) | 96.3 | 98.5 | 97.2 |
| Add. allowed (%) | 3.4 | 1.5 | 2.8 |
| RMSD bond lengths (Å) bond angles (º) | 0.004 0.086 | 0.005 1.183 | 0.006 1.291 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kou, Y.; Koag, M.-C.; Lee, S. Promutagenicity of 8-Chloroguanine, A Major Inflammation-Induced Halogenated DNA Lesion. Molecules 2019, 24, 3507. https://doi.org/10.3390/molecules24193507
Kou Y, Koag M-C, Lee S. Promutagenicity of 8-Chloroguanine, A Major Inflammation-Induced Halogenated DNA Lesion. Molecules. 2019; 24(19):3507. https://doi.org/10.3390/molecules24193507
Chicago/Turabian StyleKou, Yi, Myong-Chul Koag, and Seongmin Lee. 2019. "Promutagenicity of 8-Chloroguanine, A Major Inflammation-Induced Halogenated DNA Lesion" Molecules 24, no. 19: 3507. https://doi.org/10.3390/molecules24193507
APA StyleKou, Y., Koag, M.-C., & Lee, S. (2019). Promutagenicity of 8-Chloroguanine, A Major Inflammation-Induced Halogenated DNA Lesion. Molecules, 24(19), 3507. https://doi.org/10.3390/molecules24193507







