Characterization of Eight Novel Spiroleptosphols from Fusarium avenaceum
Abstract
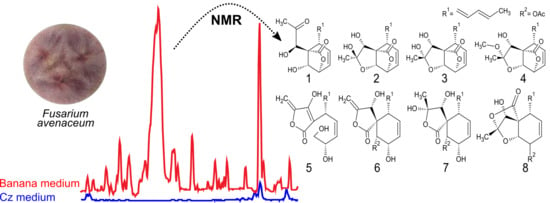
1. Introduction
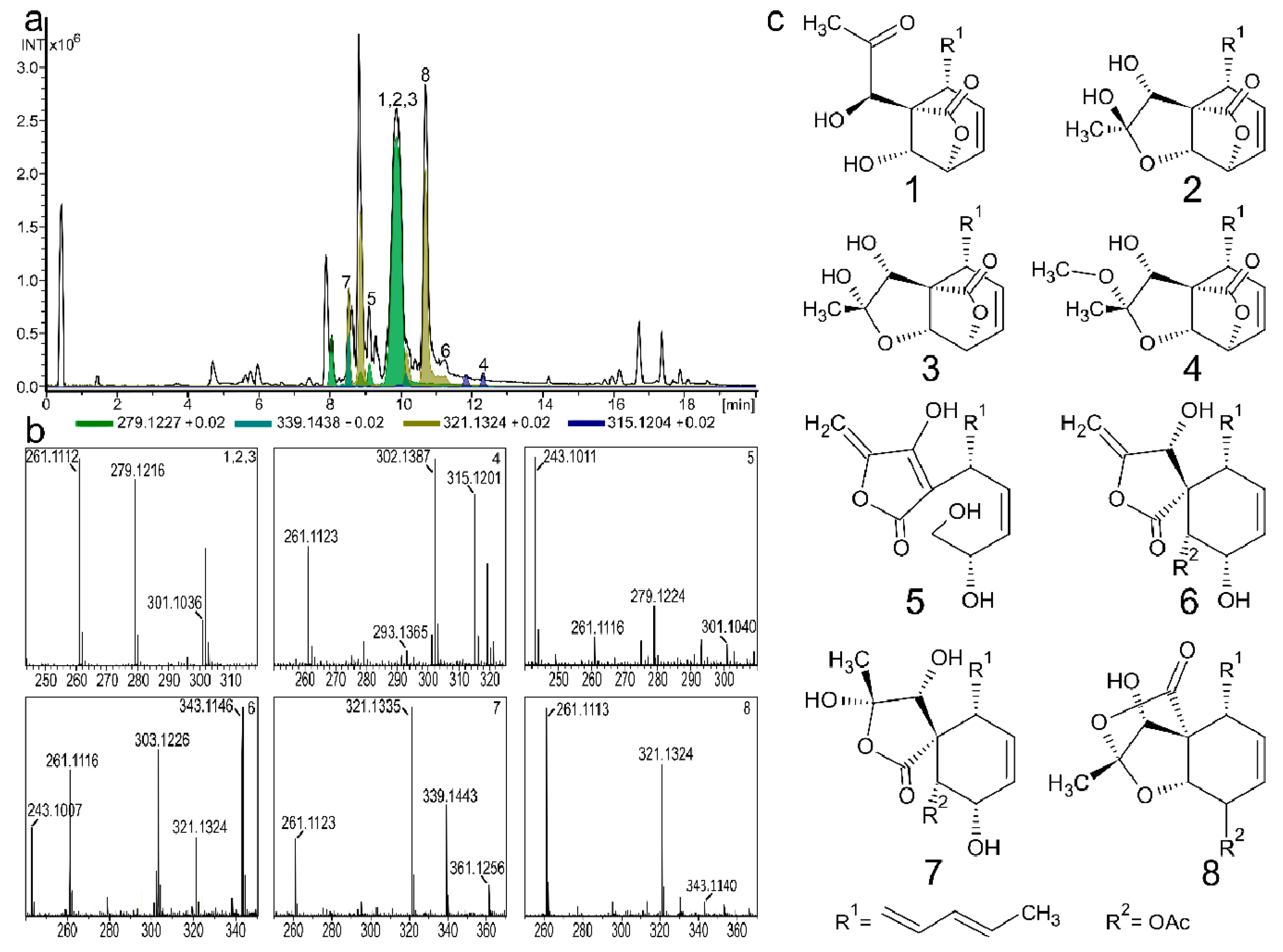
2. Results and Discussion
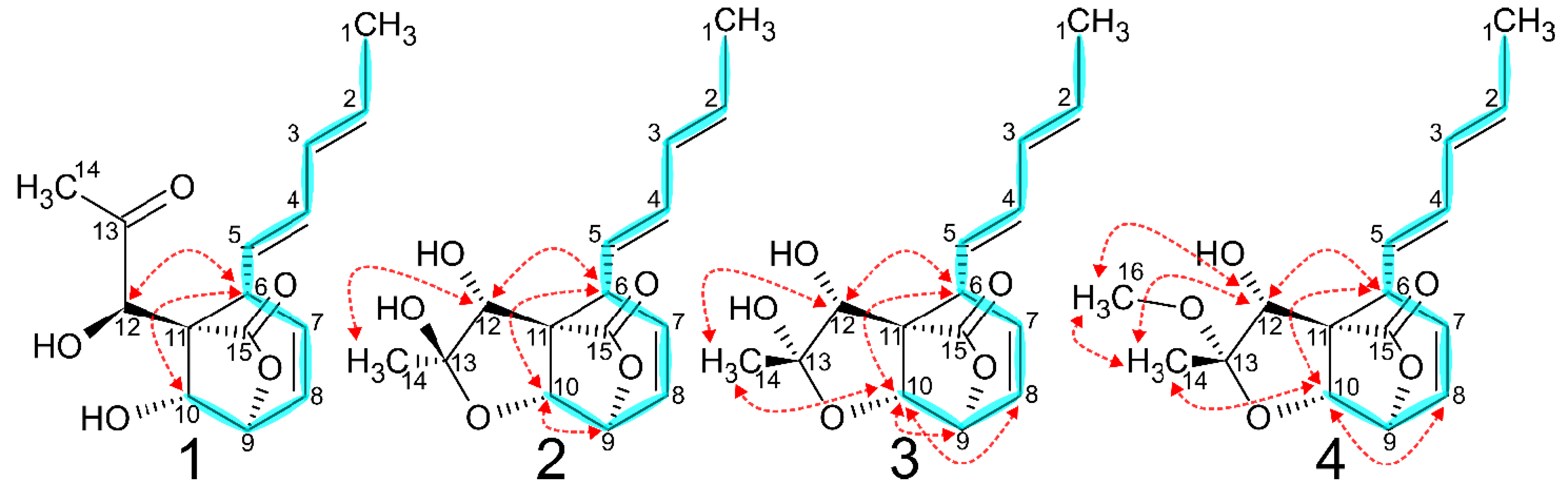
2.1. Spiroleptosphol U, T1 and T2 (1, 2 and 3)
2.2. Spiroleptosphol W (4)
2.3. Spiroleptosphol V (5)
2.4. Spiroleptosphol Z (6)
2.5. Spiroleptosphol Y (7)
2.6. Spiroleptosphol Y (8)
2.7. Absolute Configuration
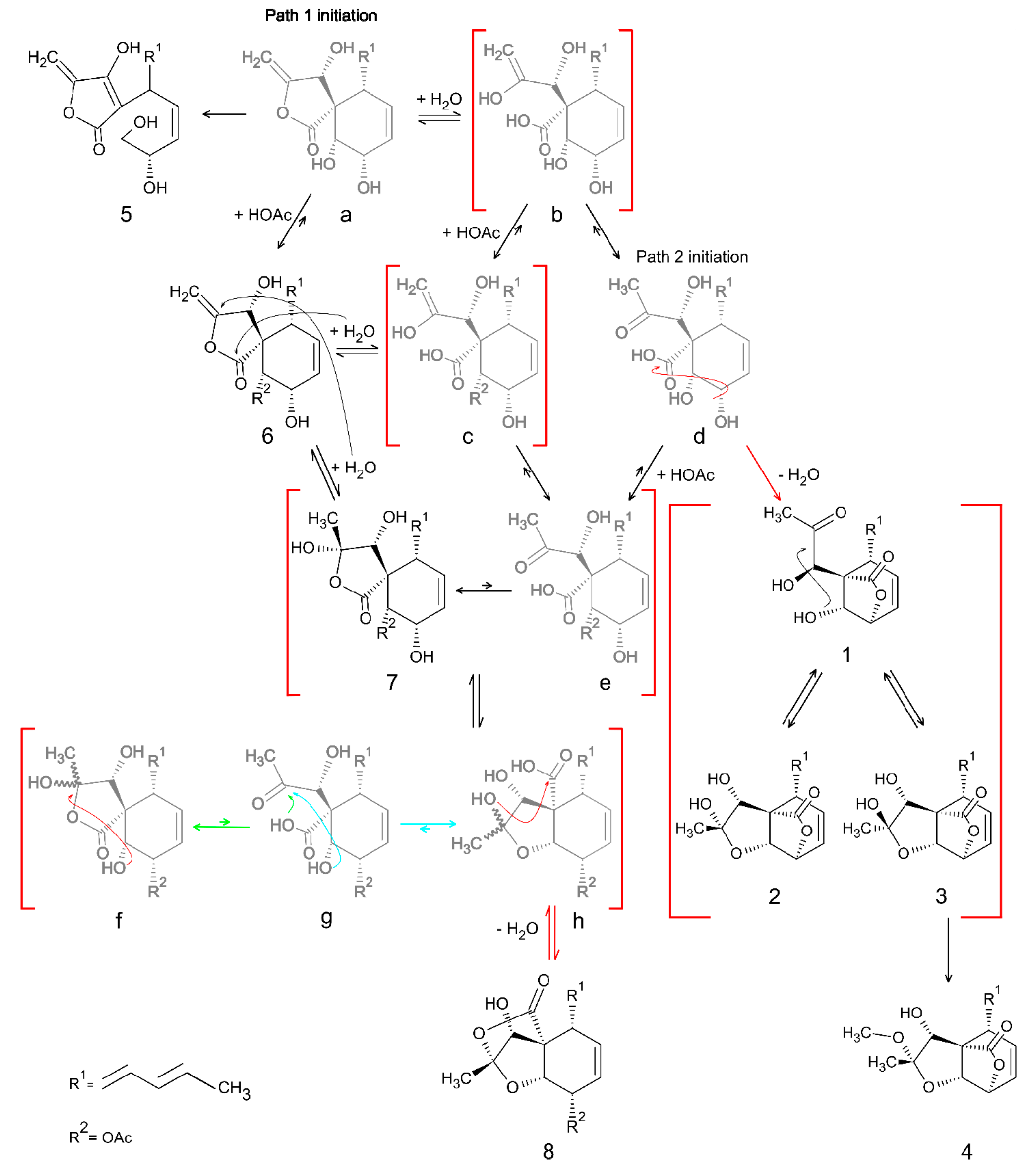
2.8. Biosynthetic Pathway
3. Materials and Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Hansen, F.T.; Gardiner, D.M.; Lysøe, E.; Fuertes, P.R.; Tudzynski, B.; Wiemann, P.; Sondergaard, T.E.; Giese, H.; Brodersen, D.E.; Sørensen, J.L. An update to polyketide synthase and non-ribosomal synthetase genes and nomenclature in Fusarium. Fungal Genet. Biol. 2015, 75, 20–29. [Google Scholar] [CrossRef]
- Brown, D.W.; Proctor, R.H. Insights into natural products biosynthesis from analysis of 490 polyketide synthases from Fusarium. Fungal Genet. Biol. 2016, 89, 37–51. [Google Scholar] [CrossRef]
- Sørensen, J.L.; Akk, E.; Thrane, U.; Giese, H.; Sondergaard, T.E. Production of fusarielins by Fusarium. Int. J. Food Microbiol. 2013, 160, 206–211. [Google Scholar] [CrossRef]
- Sondergaard, T.E.; Klitgaard, L.G.; Purup, S.; Kobayashi, H.; Giese, H.; Sørensen, J.L. Estrogenic effects of fusarielins in human breast cancer cell lines. Toxicol. Lett. 2012, 214, 259–262. [Google Scholar] [CrossRef]
- Sondergaard, T.E.; Fredborg, M.; Oppenhagen Christensen, A.M.; Damsgaard, S.K.; Kramer, N.F.; Giese, H.; Sørensen, J.L. Fast screening of antibacterial compounds from Fusaria. Toxins (Basel) 2016, 8, 355. [Google Scholar] [CrossRef]
- Westphal, K.R.; Muurmann, A.T.; Paulsen, I.E.; Nørgaard, K.T.H.; Overgaard, M.L.; Dall, S.M.; Aalborg, T.; Wimmer, R.; Sørensen, J.L.; Sondergaard, T.E. Who needs neighbors? PKS8 is a stand-alone gene in Fusarium graminearum responsible for production of gibepyrones and prolipyrone B. Molecules 2018, 23, 2232. [Google Scholar] [CrossRef]
- Bogner, C.W.; Kamdem, R.S.T.; Sichtermann, G.; Matthäus, C.; Hölscher, D.; Popp, J.; Proksch, P.; Grundler, F.M.W.; Schouten, A. Bioactive secondary metabolites with multiple activities from a fungal endophyte. Microb. Biotechnol. 2017, 10, 175–188. [Google Scholar] [CrossRef]
- Brakhage, A.A.; Schroeckh, V. Fungal secondary metabolites - Strategies to activate silent gene clusters. Fungal Genet. Biol. 2011, 48, 15–22. [Google Scholar] [CrossRef]
- Bode, H.B.; Bethe, B.; Hofs, R.; Zeeck, A. Big effects from small changes: Possible ways to explore nature’s chemical diversity. Chembiochem 2002, 3, 619–627. [Google Scholar] [CrossRef]
- Sørensen, J.L.; Sondergaard, T.E. The effects of different yeast extracts on secondary metabolite production in Fusarium. Int. J. Food Microbiol. 2014, 170, 55–60. [Google Scholar] [CrossRef]
- Sørensen, J.L.; Nielsen, K.F.; Sondergaard, T.E. Redirection of pigment biosynthesis to isocoumarins in Fusarium. Fungal Genet. Biol. 2012, 49, 613–618. [Google Scholar] [CrossRef]
- Hashimoto, M.; Tsushima, T.; Murakami, T.; Nomiya, M.; Takada, N.; Tanaka, K. Spiroleptosphol isolated from Leptosphaeria doliolum. Bioorg. Med. Chem. Lett. 2008, 18, 4228–4231. [Google Scholar] [CrossRef]
- Murakami, T.; Tsushima, T.; Takada, N.; Tanaka, K.; Nihei, K.; Miura, T.; Hashimoto, M. Four analogues of spiroleptosphol isolated from Leptosphaeria doliolum. Bioorg. Med. Chem. 2009, 17, 492–495. [Google Scholar] [CrossRef]
- Oh, H.; Swenson, D.C.; Gloer, J.B.; Shearer, C.A. New bioactive rosigenin analogues and aromatic polyketide metabolites from the freshwater aquatic fungus Massarina tunicata. J. Nat. Prod. 2003, 66, 73–79. [Google Scholar] [CrossRef]
- Albinati, A.; Brückner, S.; Camarda, L.; Nasini, G. Rosigenin, an unusual metabolite from Mycosphaerella rosigena. Tetrahedron 1980, 36, 117–121. [Google Scholar] [CrossRef]
- Wijeratne, E.M.K.; Gunaherath, G.M.K.B.; Chapla, V.M.; Tillotson, J.; de la Cruz, F.; Kang, M.; U’Ren, J.M.; Araujo, A.R.; Arnold, A.E.; Chapman, E.; et al. Oxaspirol B with p97 inhibitory activity and other oxaspirols from Lecythophora sp. FL1375 and FL1031, endolichenic fungi inhabiting Parmotrema tinctorum and Cladonia evansii. J. Nat. Prod. 2016, 79, 340–352. [Google Scholar] [CrossRef]
- Ayer, W.A.; Craw, P.A.; Neary, J. Metabolites of the fungus Arthropsis truncata. Can. J. Chem. 1992, 70, 1338–1347. [Google Scholar] [CrossRef]
- Rether, J.; Erkel, G.; Anke, T.; Sterner, O. Inhibition of inducible TNF-α expression by oxaspirodion, a novel spiro-compound from the ascomycete Chaetomium subspirale. Biol. Chem. 2004, 385, 829–834. [Google Scholar] [CrossRef]
- Namikoshi, M.; Kobayashi, H.; Yoshimoto, T.; Meguro, S.; Akano, K. Isolation and characterization of bioactive metabolites from marine-derived filamentous fungi collected from tropical and sub-tropical coral reefs. Chem. Pharm. Bull. (Tokyo) 2000, 48, 1452–1457. [Google Scholar] [CrossRef]
- Hussain, H.; Ahmed, I.; Schulz, B.; Draeger, S.; Florke, U.; Pescitelli, G.; Krohn, K. Solid-state circular dichroism and hydrogen bonding: Absolute configuration of massarigenin A from Microsphaeropsis sp. Chirality 2011, 23, 617–623. [Google Scholar] [CrossRef]
- Guiraud, P.; Steiman, R.; Seigle-Murandi, F.; Buarque De Gusmao, N. Antimicrobial and antitumor activities of mycosporulone. J. Nat. Prod. 1999, 62, 1222–1224. [Google Scholar] [CrossRef]
- Abdel-Wahab, M.A.; Asolkar, R.N.; Inderbitzin, P.; Fenical, W. Secondary metabolite chemistry of the marine-derived fungus Massarina sp., strain CNT-016. Phytochemistry 2007, 68, 1212–1218. [Google Scholar] [CrossRef]
- Fukami, A.; Taniguchi, Y.; Nakamura, T.; Rho, M.C.; Kawaguchi, K.; Hayashi, M.; Komiyama, K.; Omura, S. New members of the macrosphelides from Microsphaeropsis sp. FO-5050 IV. J. Antibiot. (Tokyo) 1999, 52, 501–504. [Google Scholar] [CrossRef]
- Murakami, T.; Takada, N.; Hashimoto, M. Biosynthetic studies of spiroleptosphol. Bioorg. Med. Chem. Lett. 2009, 19, 1122–1125. [Google Scholar] [CrossRef]
- Sørensen, J.L.; Giese, H. Influence of carbohydrates on secondary metabolism in Fusarium avenaceum. Toxins (Basel) 2013, 5, 1655–1663. [Google Scholar] [CrossRef]
- Robien, W. A Critical evaluation of the quality of published 13C NMR data in natural product chemistry. Prog. Chem. Org. Nat. Prod. 2017, 105, 137–215. [Google Scholar] [CrossRef]
- Perdew, J.P.; Burke, K.; Wang, Y. Generalized gradient approximation for the exchange-correlation hole of a many-electron system. Phys. Rev. B Condens. Matter 1996, 54, 16533–16539. [Google Scholar] [CrossRef]
- Klamt, A.; Schüürmann, G. COSMO: A new approach to dielectric screening in solvents with explicit expressions for the screening energy and its gradient. J. Chem. Soc. Perkins Trans. 2 1993, 0, 799–805. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian09; Revision D01; Gaussian Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
Sample Availability: Samples of the compounds can be purified from fungal strains. Strains are available from the authors. |



| Spiroleptosphol U (1) | Spiroleptosphol T1 (2) | |||||||
| # | δC | type | δH (J in Hz) | HMBC 2 | δC | type | δH (J in Hz) | HMBC 2 |
| 1 | 18.2 | CH3 | 1.74 1 | 2–4,6,7,11 | 18.2 | CH3 | 1.74 1 | 2–6,7,11 |
| 2 | 131.5 | CH | 5.71 1 | 1,4–6 | 130.7 | CH | 5.70 1 | 1,4,5 |
| 3 | 131.9 | CH | 6.03 1 | 1,4–6 | 132.3 | CH | 6.06 1 | 1,4,5 |
| 4 | 136.6 | CH | 6.05 1 | 1–3,6 | 134.7 | CH | 6.10 1 | 2,3,5,6,11 |
| 5 | 127.5 | CH | 5.24 (ddd, 1.3;9.6;14.4) | 1–3,6,7,11 | 128.1 | CH | 5.51 (dd, 7.9;14.9) | 2,3,6,7,11 |
| 6 | 46.5 | CH | 3.64 (ddd, 2.7;9.6) | 4,7–12,15 | 47.9 | CH | 3.44 1 | 4,5,7–12,15 |
| 7 | 135.4 | CH | 5.61 (ddd, 1.0;2.9;9.2) | 5,6,9–12 | 136.1 | CH | 5.86 (dd, 2.6;9.4) | 5,6,8–12 |
| 8 | 127.6 | CH | 6.16 (ddd, 2.7;5.8;9.2) | 5–7,9–11 | 128.3 | CH | 6.25 (ddd, 2.6;5.9;9.4) | 4–7,9–11 |
| 9 | 79.2 | CH | 4.52 (d, 5.8) | 7,8,10–12,15 | 75.2 | CH | 4.75 (dd, 5.9) | 6–8,10–13,15 |
| 10 | 78.6 | CH | 4.51 (s) | 6,11–13,15 | 82.7 | CH | 4.41 (s) | 6,8,9,11,12,14,15 |
| 11 | 59.0 | C | 59.0 | C | ||||
| 12 | 75.7 | CH | 4.36 (s) | 6,10,11,13,15 | 86.0 | CH | 4.33 (s) | 6,11,13–15 |
| 13 | 212.6 | C | 109.6 | C | ||||
| 14 | 28.4 | CH3 | 2.35 (s) | 12 | 24.0 | CH3 | 1.34 (s) | 6,11,12,13 |
| 15 | 176.0 | C | 177.9 | C | ||||
| Spiroleptosphol T2 (3) | Spiroleptosphol W (4) | |||||||
| # | δC | type | δH (J in Hz) | HMBC 2 | δC | type | δH (J in Hz) | HMBC 2 |
| 1 | 18.2 | CH3 | 1.74 1 | 2,4–6,11 | 18.2 | CH3 | 1.74 (dd, 1.3;6.8) | 2–5 |
| 2 | 130.3 | CH | 5.69 1 | 1,4,5 | 130.7 | CH | 5.70 (dq, 6.7;14.6) | 1,4,5 |
| 3 | 132.5 | CH | 6.06 1 | 1,3,5,6,11 | 132.3 | CH | 6.05 (ddq, 1.5;10.5;14.7) | 1,4,5 |
| 4 | 134.4 | CH | 6.09 1 | 2,6 | 134.7 | CH | 6.11 (dd, 10.4;15.0) | 2,3,5–7,11 |
| 5 | 128.5 | CH | 5.59 1 | 3,6,7,11 | 128.1 | CH | 5.50 (dd, 7.9;14.9) | 2,3,6,7,11 |
| 6 | 48.2 | CH | 3.43 1 | 4,5,7–12,15 | 48.0 | CH | 3.44 (ddd, 2.5;2.5;8.0) | 4,5,7,11,12,15 |
| 7 | 136.0 | CH | 5.82 (dd, 2.5;9.3) | 5,6,8–12 | 136.2 | CH | 5.85 (ddd, 0.6;2.7;9.4) | 5,6,9–12 |
| 8 | 128.8 | CH | 6.23 (ddd, 2.7;5.8;9.5) | 4–7,9–11 | 128.2 | CH | 6.25 | 5–7,9–11 |
| 9 | 75.7 | CH | 4.73 (d, 5.8) | 6–8,10–12,15 | 75.0 | CH | 4.79 (dd, 0.6;5.9) | 6–8,10,11,15 |
| 10 | 83.5 | CH | 4.24 (s) | 6,8,9,12–15 | 83.0 | CH | 4.31 (s) | 6,8,9,11,12,15 |
| 11 | 57.1 | C | 58.9 | C | ||||
| 12 | 81.9 | CH | 4.17 (s) | 6,11,13–15 | 86.0 | CH | 4.32 (s) | 6,11,13–15 |
| 13 | 104.7 | C | 112.8 | C | ||||
| 14 | 25.9 | CH3 | 1.48 (s) | 11–13 | 18.9 | CH3 | 1.30 (s) | 12,13 |
| 15 | 177.2 | C | 177.8 | C | ||||
| 16 | 49.6 | CH3 | 3.30 (s) | 12,13 | ||||
| Spiroleptosphol V (5) | Spiroleptosphol Z (6) | |||||||
| # | δC | type | δH (J in Hz) | HMBC 2 | δC | type | δH (J in Hz) | HMBC 2 |
| 1 | 17.9 | CH3 | 1.71 (d, 7.3) | 2–5 | 18.0 | CH3 | 1.73 (dd, 1.4;6.8) | 2–6,11 |
| 2 | 128.8 | CH | 5.62 (dq, 6.9;14.2) | 1,4–6 | 130.7 | CH | 5.66 (dq, 6.7;15.0) | 1,3–5 |
| 3 | 132.4 | CH | 6.03 1 | 1,4,5 | 132.0 | CH | 5.95 (ddq, 1.5;10.3;15.0) | 1,4,5 |
| 4 | 131.5 | CH | 6.05 1 | 2,3,6 | 135.5 | CH | 6.06 (dd, 10.3;15.2) | 1–3,5,6,11 |
| 5 | 131.0 | CH | 5.71 (dd, 6.9;14.5) | 1–3,6,11 | 128.6 | CH | 5.51 (dd, 8.7;15.2) | 2–4,6,7,11 |
| 6 | 36.2 | CH | 4.30 (dd, 7.1;9.6) | 5,7,8,11,12,15 | 40.1 | CH | 3.43 (ddd, 2.1:2.4;8.7) | 4,5,8,10–12,15 |
| 7 | 132.5 | CH | 5.89 (ddd, 1.1;9.8;11.0) | 4,6,9,10,11 | 130.3 | CH | 5.63 (ddd, 1.7;2.8;10.2) | 5,6,9–11 |
| 8 | 129.9 | CH | 5.36 (ddd, 1.0;8.5;11.0) | 5,6,9,10,11 | 128.3 | CH | 5.82 (ddd, 2.6;3.8;10.2) | 3,5–7,9–11 |
| 9 | 69.4 | CH | 4.56 (dddd, 1.2;4.5,7.0,8.3) | 7,8,10 | 64.3 | CH | 4.39 (ddd, 1.8;3.6;6.9) | 6–8,10,11,15 |
| 10 | 66.9 | CH2 | 3.40 (dd, 7.0;11.2) | 8,9 | 72.0 | CH | 5.16 (d, 5.0) | 6–9,11,12,15,16 |
| 3.44 (dd, 4.5;11.2) | 8,9 | |||||||
| 11 | 103.9 | C | 55.2 | C | ||||
| 12 | 166.7 | C | 70.9 | CH | 4.94 (dd, 2.3;2.6) | 6,10,11,13–15 | ||
| 13 | 153.0 | C | 158.9 | C | ||||
| 14 | 90.6 | CH2 | 4.91 (d, 2.6) | 6,11–13 | 89.2 | CH2 | 4.76 (dd, 2.6;2.6) | 10–13,15 |
| 5.06 (d, 2.6) | 6,12,13,15 | 4.63 (dd, 2.3;2.6) | 10–13,15 | |||||
| 15 | 172.3 | C | 174.6 | C | ||||
| 16 | 171.8 | C | ||||||
| 17 | 20.7 | CH3 | 2.12 (s) | 9,10,16 | ||||
| Spiroleptosphol Y (7) | Spiroleptosphol X (8) | |||||||
| # | δC | type | δH (J in Hz) | HMBC 2 | δC | type | δH (J in Hz) | HMBC 2 |
| 1 | 18.0 | CH3 | 1.71 (d, 6.5) | 2,3 | 17.9 | CH3 | 1.74 (d, 6.7) | 2–5 |
| 2 | 129.0 | CH | 5.60 1 | 1 | 129.3 | CH | 5.67 (dq, 6.8;14.8) | 1,3–5 |
| 3 | 132.7 | CH | 5.99 1 | 1 | 132.4 | CH | 6.06 (dd, 14.8;10.5) | 4,7 |
| 4 | 133.1 | CH | 6.00 1 | 6 | 134.1 | CH | 6.21 (dd, 10.4;15.3) | 2,3,6,7,11 |
| 5 | 131.9 | CH | 5.64 1 | 1,3 | 128.5 | CH | 5.96 (dd, 8.5;15.3) | 2–4,6,7,11 |
| 6 | 40.3 | CH | 3.65 1 | 4,5 | 41.6 | CH | 3.32 1 | |
| 7 | 131.5 | CH | 5.59 1 | 6,9,11 | 137.4 | CH | 5.91 (d, 10.0) | 5,6,8,9,11 |
| 8 | 127.6 | CH | 5.58 1 | 6,9,11 | 122.8 | CH | 5.74 1 | 5–7,9–11 |
| 9 | 65.5 | CH | 4.39 1 | 63.9 | CH | 5.42 (dd, 4.6;4.6) | 7,8,10,11,16 | |
| 10 | 73.3 | CH | 5.31 (d, 3.8) | 6,8,9,11,15,16 | 77.0 | CH | 4.38 (d, 4.3) | 6,9,11–13,15,17 |
| 11 | 52.5 | C | 56.5 | C | ||||
| 12 | 77.5 | CH | 4.01 (s) | 6,10,11,14 | 83.8 | CH | 4.23 (s) | 6,10,13,15 |
| 13 | 104.7 | C | 111.9 | C | ||||
| 14 | 25.0 | CH3 | 1.60 (s) | 12,13 | 14.8 | CH3 | 1.55 (s) | 6,10–13,15 |
| 15 | 175.9 | C | 174.6 | C | ||||
| 16 | 172.6 | C | 171.7 | C | ||||
| 17 | 21.2 | CH3 | 2.10 (s) | 16 | 20.4 | CH3 | 2.01 (s) | 9,16 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Westphal, K.R.; Werner, M.I.H.; Nielsen, K.A.H.; Sørensen, J.L.; Andrushchenko, V.; Winde, J.; Hertz, M.; Jensen, M.A.; Mortensen, M.L.; Bouř, P.; et al. Characterization of Eight Novel Spiroleptosphols from Fusarium avenaceum. Molecules 2019, 24, 3498. https://doi.org/10.3390/molecules24193498
Westphal KR, Werner MIH, Nielsen KAH, Sørensen JL, Andrushchenko V, Winde J, Hertz M, Jensen MA, Mortensen ML, Bouř P, et al. Characterization of Eight Novel Spiroleptosphols from Fusarium avenaceum. Molecules. 2019; 24(19):3498. https://doi.org/10.3390/molecules24193498
Chicago/Turabian StyleWestphal, Klaus Ringsborg, Manuela Ilse Helga Werner, Katrine Amalie Hamborg Nielsen, Jens Laurids Sørensen, Valery Andrushchenko, Jacob Winde, Morten Hertz, Mikkel Astrup Jensen, Mathilde Lauge Mortensen, Petr Bouř, and et al. 2019. "Characterization of Eight Novel Spiroleptosphols from Fusarium avenaceum" Molecules 24, no. 19: 3498. https://doi.org/10.3390/molecules24193498
APA StyleWestphal, K. R., Werner, M. I. H., Nielsen, K. A. H., Sørensen, J. L., Andrushchenko, V., Winde, J., Hertz, M., Jensen, M. A., Mortensen, M. L., Bouř, P., Sondergaard, T. E., & Wimmer, R. (2019). Characterization of Eight Novel Spiroleptosphols from Fusarium avenaceum. Molecules, 24(19), 3498. https://doi.org/10.3390/molecules24193498