Cytotoxic Polyketides from a Deep-Sea Sediment Derived Fungus Diaporthe phaseolorum FS431
Abstract
:1. Introduction
2. Results and Discussion
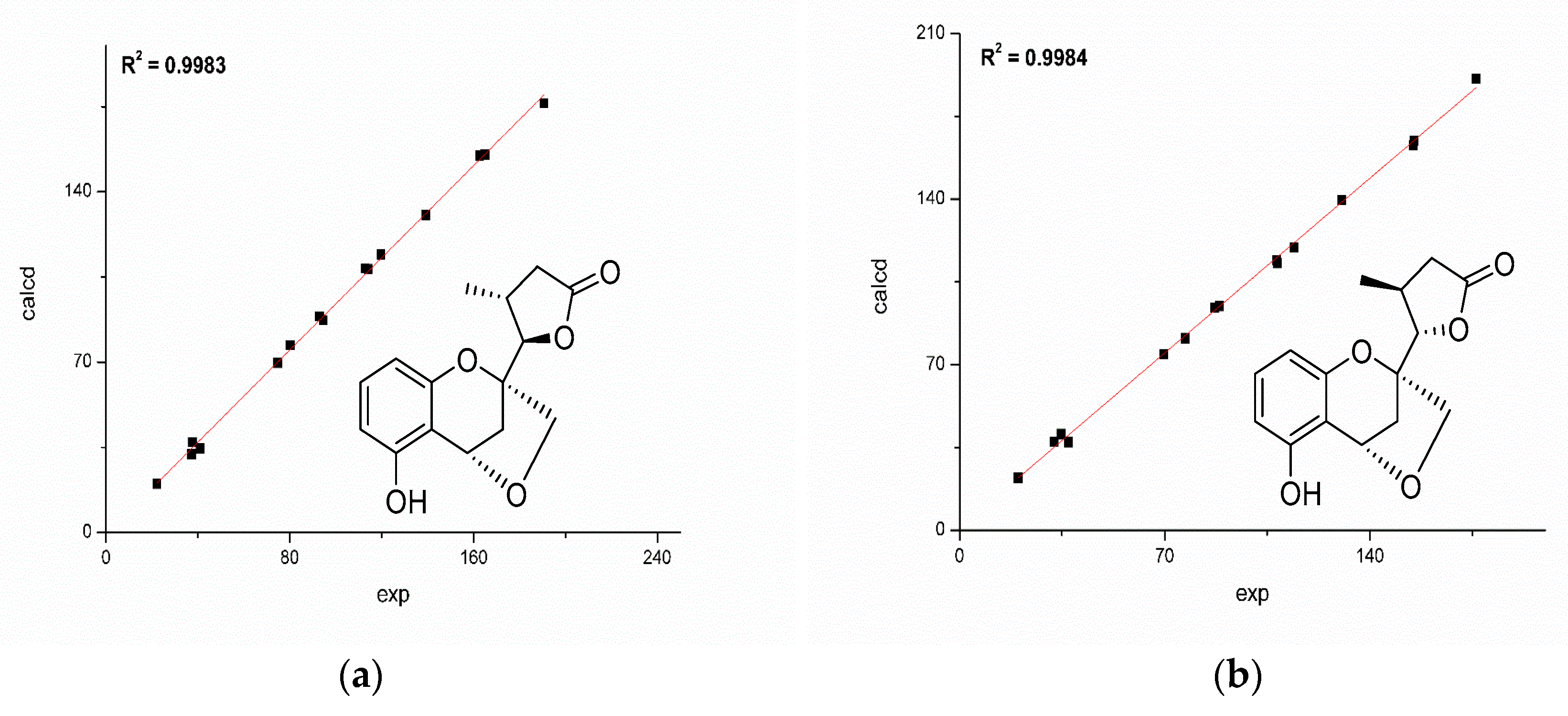
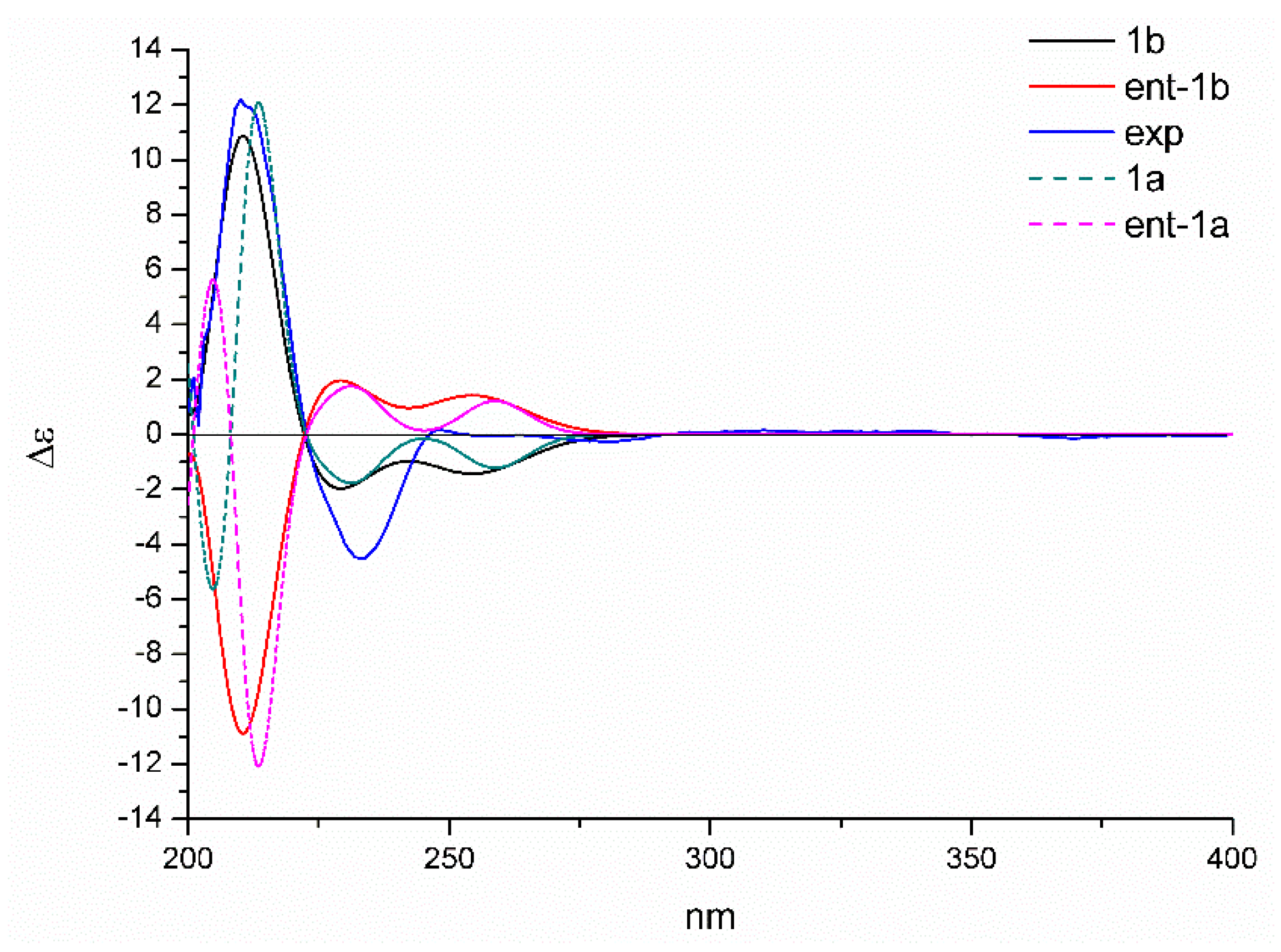
2.1. Structure Elucidation
2.2. Cytotoxicity Activity
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Fungal Material
3.3. Fermentation and Extraction
3.4. Isolation and Purification
3.5. Quantum Chemical Calculation of NMR Chemical Shifts and ECD Spectra
3.6. Cytotoxic Activity Assay
3.7. Spectroscopic Data
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Bugni, T.S.; Ireland, C.M. Marine-derived fungi: A chemically and biologically diverse group of microorganisms. Nat. Prod. Rep. 2004, 21, 143–163. [Google Scholar] [CrossRef] [PubMed]
- Rateb, M.E.; Ebel, R. Secondary metabolites of fungi from marine habitats. Nat. Prod. Rep. 2011, 28, 290. [Google Scholar] [CrossRef] [PubMed]
- Skropeta, D.; Wei, L. Recent advances in deep-sea natural products. Nat. Prod. Rep. 2014, 31, 999–1025. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Liao, Y.; Zhang, B.; Gao, M.; Ke, W.; Li, F.; Shao, Z. Citrinin Monomer and Dimer Derivatives with Antibacterial and Cytotoxic Activities Isolated from the Deep Sea-Derived Fungus Penicillium citrinum NLG-S01-P1. Mar. Drugs 2019, 17, 46. [Google Scholar] [CrossRef] [PubMed]
- Pang, X.; Lin, X.; Yang, J.; Zhou, X.; Yang, B.; Wang, J.; Liu, Y. Spiro-Phthalides and Isocoumarins Isolated from the Marine-Sponge-Derived Fungus Setosphaeria sp. SCSIO41009. J. Nat. Prod. 2018, 81, 1860–1868. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.W.; Yang, J.; Chen, F.M.; Lin, X.P.; Chen, C.M.; Zhou, X.F.; Liu, S.W.; Liu, Y.H. Structurally diverse polyketides from the mangrove-derived fungus Diaporthe sp. SCOSIC 41011 with their anti-influenza A virus activities. Front. Chem. 2018, 6, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Chang, F.-R.; Wang, S.-W.; Li, C.-Y.; Lu, Y.-Y.; Liu, S.-Y.V.; Chen, C.-Y.; Wu, Y.-C.; Cheng, Y.-B. Natural Products from Diaporthe arecae with Anti-Angiogenic Activity. Isr. J. Chem. 2019, 59, 439–445. [Google Scholar] [CrossRef]
- Li, G.; Kusari, S.; Kusari, P.; Kayser, O.; Spiteller, M. Endophytic Diaporthe sp. LG23 produces a potent antibacterial tetracyclic triterpenoid. J. Nat. Prod. 2015, 78, 2128–2132. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.; Liao, Z.; Zhou, M.; Wang, G.; Wu, Y.; Gao, S.; Qiu, D.; Liu, X.; Lin, T.; Chen, H. Cytoskyrin C, an unusual asymmetric bisanthraquinone with cage-like skeleton from the endophytic fungus Diaporthe sp. Fitoterapia 2018, 128, 253–257. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.W.; Ondeyka, J.G.; Herath, K.B.; Guan, Z.Q.; Collado, J.; Platas, G.; Pelaez, F.; Leavitt, P.S.; Gurnett, A.; Nare, B.; et al. Tenellones A and B from a Diaporthe sp.: Two highly substituted benzophenone inhibitors of parasite cGMP-dependent protein kinase activity. J. Nat. Prod. 2005, 68, 611–613. [Google Scholar] [CrossRef] [PubMed]
- Brady, S.F.; Wagenaar, M.M.; Singh, M.P.; Janso, J.E.; Clardy, J. The Cytosporones, New Octaketide Antibiotics Isolated from an Endophytic Fungus. Org. Lett. 2000, 2, 4043–4046. [Google Scholar] [CrossRef] [PubMed]
- Calcul, L.; Waterman, C.; Ma, W.S.; Lebar, M.D.; Harter, C.; Mutka, T.; Morton, L.; Maignan, P.; Olphen, A.; Kyle, D.E.; et al. Screening Mangrove Endophytic Fungi for Antimalarial Natural Products. Mar. Drugs 2013, 11, 5036–5050. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Yang, X.Q.; Wan, C.P.; Wang, B.Y.; Yin, H.Y.; Shi, L.J.; Wu, Y.M.; Yang, Y.B.; Zhou, H.; Ding, Z.T. Potential antihyperlipidemic polyketones from endophytic Diaporthe sp. JC-J7 in Dendrobium nobile. RSC Adv. 2018, 8, 41810–41817. [Google Scholar] [CrossRef] [Green Version]
- Isaka, M.; Jaturapat, A.; Rukseree, K.; Danwisetkanjana, K.; Tanticharoen, M.; Thebtaranonth, Y. Phomoxanthones A and B, Novel Xanthone Dimers from the Endophytic FungusPhomopsisSpecies. J. Nat. Prod. 2001, 64, 1015–1018. [Google Scholar] [CrossRef] [PubMed]
- Wagenaar, M.M.; Clardy, J. Dicerandrols, new antibiotic and cytotoxic dimers produced by the fungus Phomopsis longicolla isolated from an endangered mint. J. Nat. Prod. 2001, 64, 1006–1009. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.-H.; He, F.-J.; Hua, H.-M.; Liu, B.; Wang, H.-F.; Liu, F.; Bai, J.; Chen, G.; Pei, Y.-H. Isolation of a New Compound from Penicillium oxalicum. Chem. Nat. Compd. 2016, 52, 821–823. [Google Scholar]
- Guo, H.; Liu, Z.-M.; Chen, Y.-C.; Tan, H.-B.; Li, S.-N.; Li, H.-H.; Gao, X.-X.; Liu, H.-X.; Zhang, W.-M. Chromone-Derived Polyketides from the Deep-Sea Fungus Diaporthe phaseolorum FS431. Mar. Drugs 2019, 17, 182. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Liu, Y.N.; Li, J.; Huang, X.S.; Yan, T.; Cao, W.H.; Liu, H.J.; Long, Y.H.; She, Z.G. Diaporindenes A−D: Four unusual 2,3-dihydro-1H-indene analogues with anti-inflammatory activities from the mangrove endophytic fungus Diaporthe sp. SYSU-HQ3. J. Org. Chem. 2018, 83, 11804–11813. [Google Scholar] [CrossRef] [PubMed]
- Skehan, P.; Scudiero, D.; Vistica, D.; Bokesch, H.; Kenney, S.; Storeng, R.; Monks, A.; McMahon, J.; Warren, J.T.; Boyd, M.R. New Colorimetric Cytotoxicity Assay for Anticancer-Drug Screening. J. Natl. Cancer Inst. 1990, 82, 1107–1112. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds 1–6 are available from the authors. |
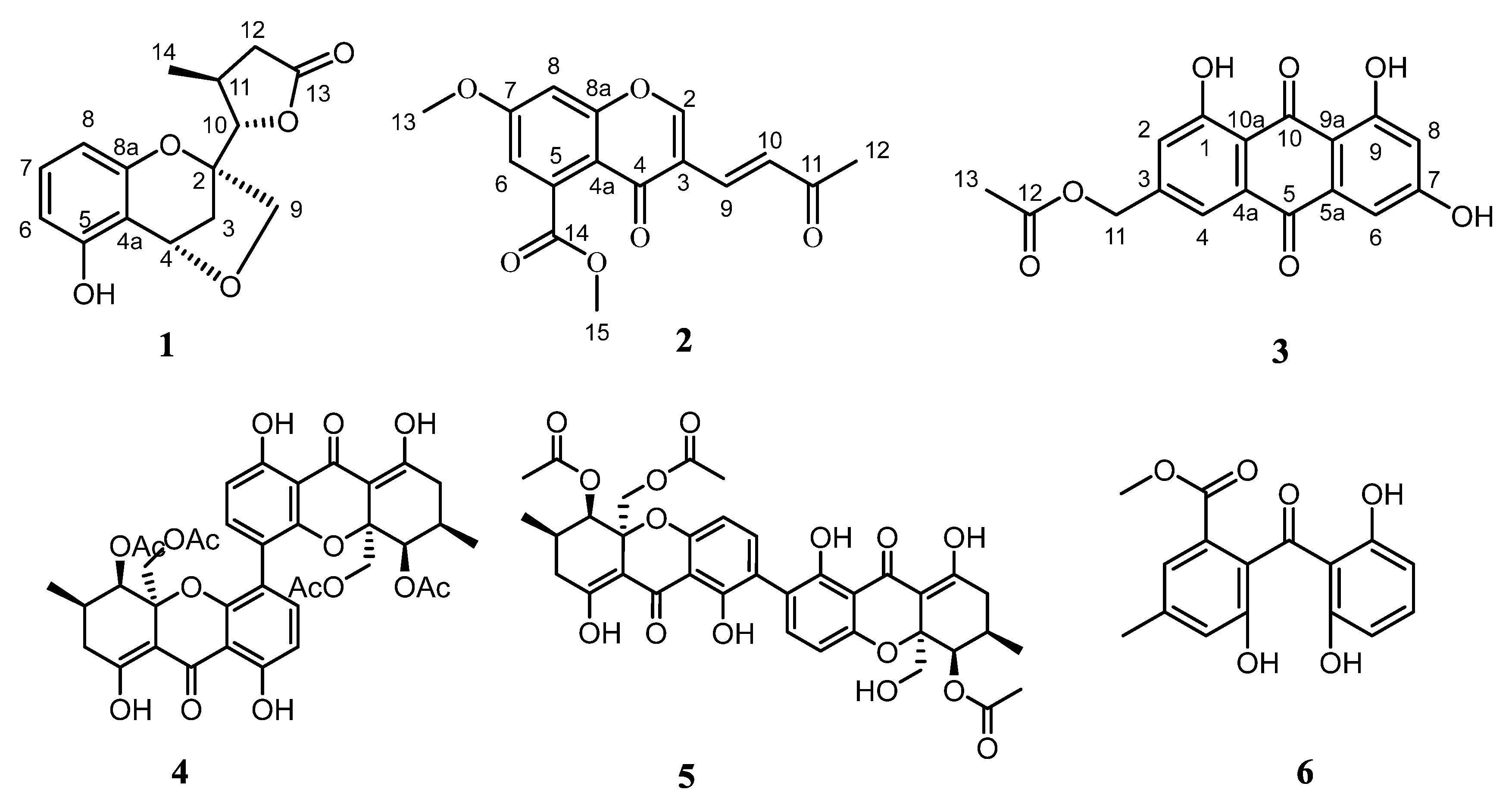
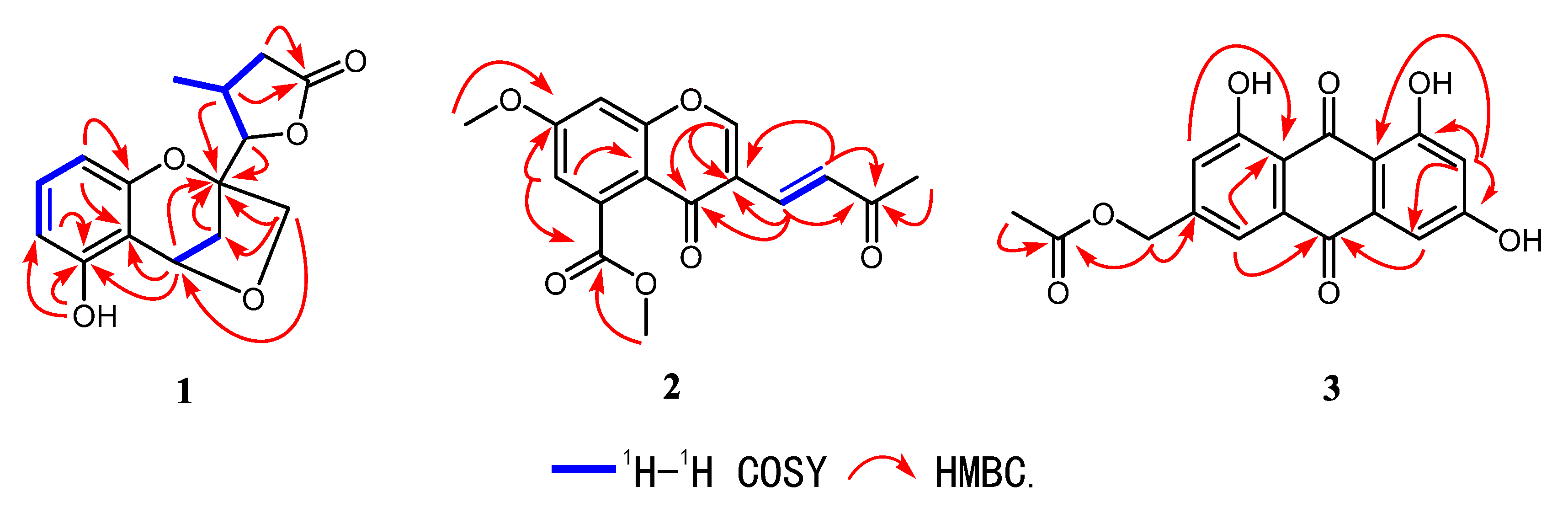

| No. | δH (J in Hz) | δC |
|---|---|---|
| 2 | 87.2, C | |
| 3a | 2.22 (1H, m) | 34.9, CH2 |
| 3b | 2.14 (1H, m) | |
| 4 | 5.44 (1H, d, 5.0) | 69.7, CH |
| 4a | 114.2, C | |
| 5 | 154.8, C | |
| 6 | 6.42 (1H, br d, 8.0) | 108.5, CH |
| 7 | 6.97 (1H, t, 8.1) | 130.4, CH |
| 8 | 6.31 (1H, br d, 8.0) | 108.2, CH |
| 8a | 155.2, C | |
| 9 | 4.05 (2H, s) | 77.0, CH2 |
| 10 | 4.51 (1H, d, 4.0) | 88.7, CH |
| 11 | 2.08 (1H, m) | 32.2, CH |
| 12a | 2.24 (1H, m) | 37.4, CH2 |
| 12b | 2.85 (1H, overlapped) | |
| 13 | 176.4, C | |
| 14 | 1.28 (3H, d, 6.1) | 20.0, CH3 |
| 5-OH | 8.59 (1H, br s) |
| No. | δH (J in Hz) | δC |
|---|---|---|
| 2 | 8.08 (1H, s) | 156.5, CH |
| 3 | 119.6, C | |
| 4 | 174.2, C | |
| 4a | 114.8, C | |
| 5 | 135.3, C | |
| 6 | 6.96 (1H, d, 2.4) | 113.9, CH |
| 7 | 163.2, C | |
| 8 | 6.92 (1H, d, 2.4) | 101.8, CH |
| 8a | 157.3, C | |
| 9 | 7.29 (1H, d, 16.1) | 133.0, CH |
| 10 | 7.41 (1H, d, 16.1) | 129.7, CH |
| 11 | 198.7, C | |
| 12 | 2.35 (3H, s) | 28.6, CH3 |
| 13 | 3.93 (3H, s) | 56.2, CH3 |
| 14 | 169.2, C | |
| 15 | 4.03 (3H, s) | 53.2, CH3 |
| Position | δH (J in Hz) | δC |
|---|---|---|
| 1 | 161.3, C | |
| 2 | 7.30 (1H, br s) | 122.3, CH |
| 3 | 145.6, C | |
| 4 | 7.63 (1H, br s) | 117.8, CH |
| 4a | 133.3, C | |
| 5 | 181.3, C | |
| 5a | 135.1, C | |
| 6 | 7.12 (1H, br d, 2.4) | 109.2, CH |
| 7 | 166.2, C | |
| 8 | 6.58 (1H, br d, 2.4) | 108.0, CH |
| 9 | 164.6, C | |
| 9a | 108.9, C | |
| 10 | 189.4, C | |
| 10a | 115.1, C | |
| 11 | 5.18 (2H, s) | 64.2, CH2 |
| 12 | 170.2, C | |
| 13 | 2.14 (3H, s) | 20.6, CH3 |
| 1-OH | 12.12 (1H, br s) | 161.3, C |
| 9-OH | 12.12 (1H, br s) |
| NO. | SF-268 | MCF-7 | HepG-2 | A549 |
|---|---|---|---|---|
| 4 | 37.86 ± 1.28 | 2.60 ± 0.28 | 2.55 ± 0.06 | 4.64 ± 0.30 |
| 5 | 45.26 ± 3.18 | 48.08 ± 1.55 | 49.58 ± 0.45 | 38.64 ± 1.42 |
| doxorubicin | 0.57 ± 0.04 | 0.95 ± 0.06 | 1.18 ± 0.15 | 0.70 ± 0.04 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niu, Z.; Chen, Y.; Guo, H.; Li, S.-N.; Li, H.-H.; Liu, H.-X.; Liu, Z.; Zhang, W. Cytotoxic Polyketides from a Deep-Sea Sediment Derived Fungus Diaporthe phaseolorum FS431. Molecules 2019, 24, 3062. https://doi.org/10.3390/molecules24173062
Niu Z, Chen Y, Guo H, Li S-N, Li H-H, Liu H-X, Liu Z, Zhang W. Cytotoxic Polyketides from a Deep-Sea Sediment Derived Fungus Diaporthe phaseolorum FS431. Molecules. 2019; 24(17):3062. https://doi.org/10.3390/molecules24173062
Chicago/Turabian StyleNiu, Zheng, Yuchan Chen, Heng Guo, Sai-Ni Li, Hao-Hua Li, Hong-Xin Liu, Zhaoming Liu, and Weimin Zhang. 2019. "Cytotoxic Polyketides from a Deep-Sea Sediment Derived Fungus Diaporthe phaseolorum FS431" Molecules 24, no. 17: 3062. https://doi.org/10.3390/molecules24173062





