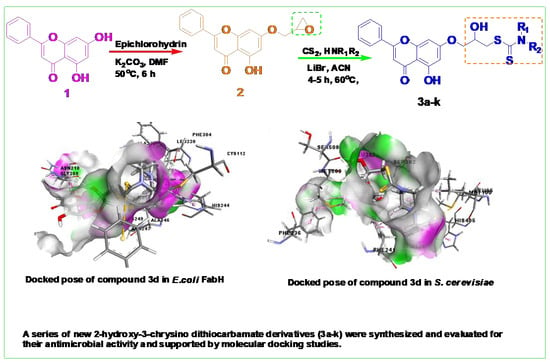
Molecular Design, Synthesis, and Biological Evaluation of 2-Hydroxy-3-Chrysino Dithiocarbamate Derivatives
Abstract
1. Introduction
2. Results
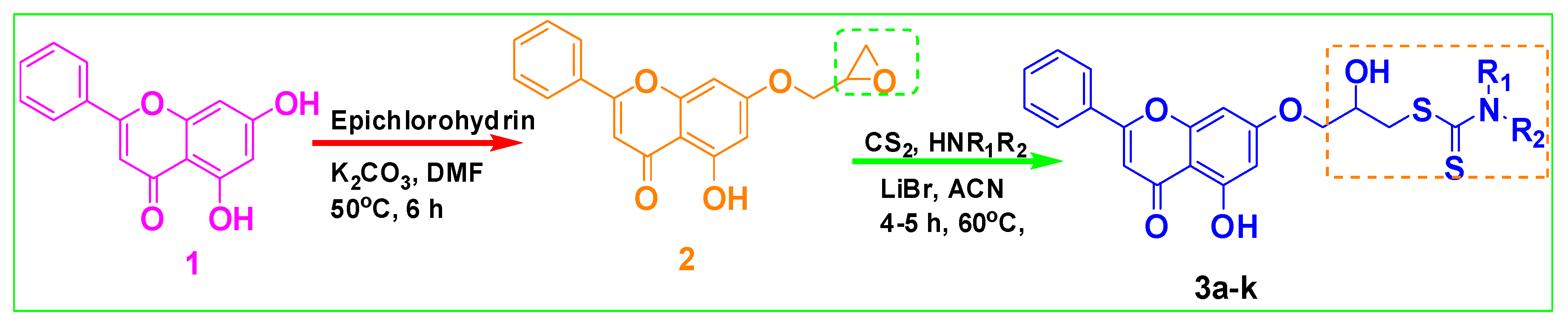
2.1. Chemistry
2.2. Pharmacology
2.2.1. Antimicrobial Evaluation
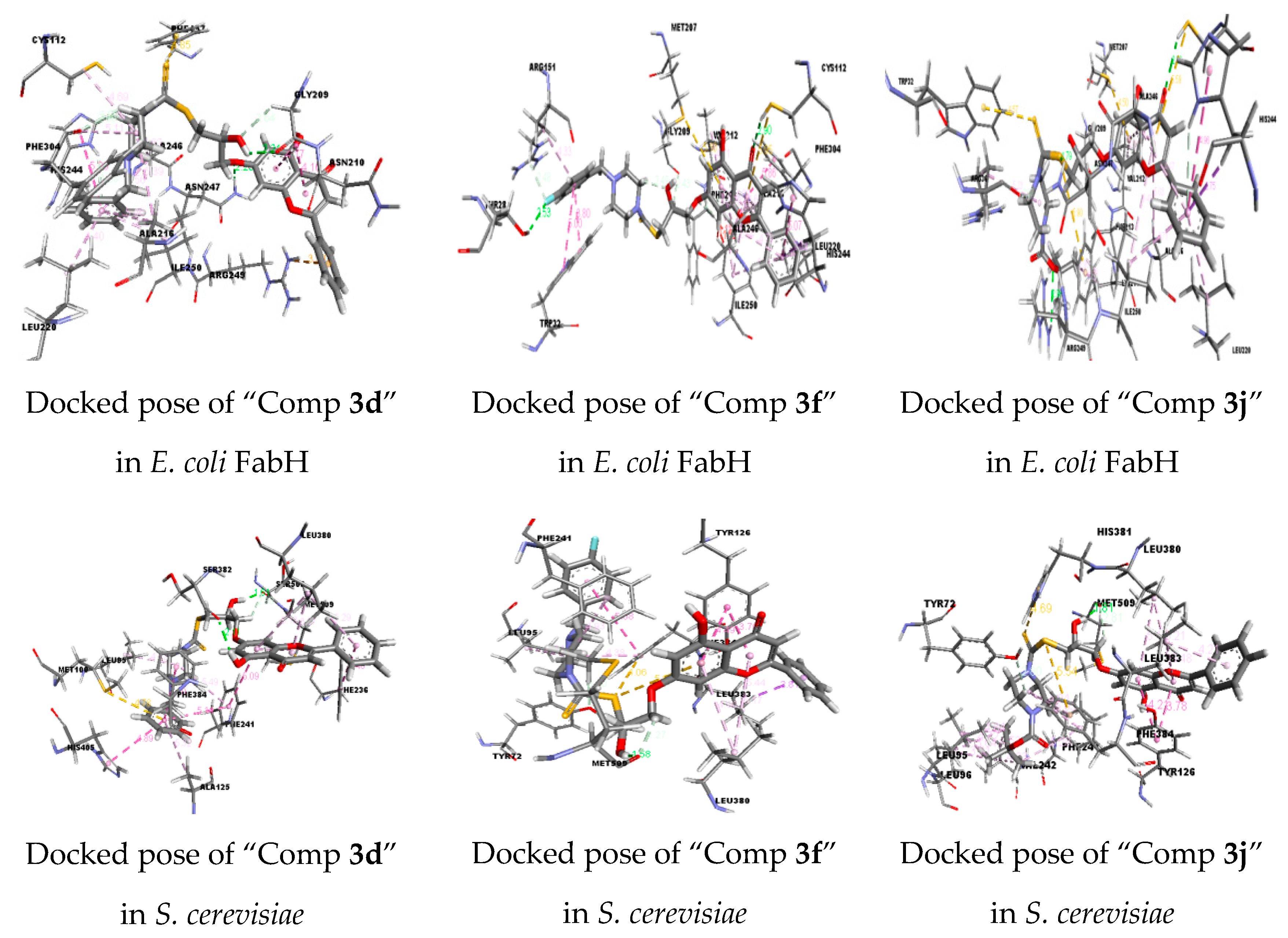
2.2.2. Molecular Docking Studies
2.2.3. Adsorption, Distribution, Metabolism, and Excretion (ADME)-Profile
3. Materials and Methods
3.1. Chemistry
3.1.1. General
3.1.2. Synthesis of 5-hydroxy-7-(oxiran-2-ylmethoxy)-2-phenyl-4H-chromen-4-one (2):
3.1.3. Typical Procedure for the Synthesis of 3a–k
3.2. Preparation of the Protein and the Ligand for Docking Simulations
3.3. Docking Simulation of the Synthesized Compounds
3.4. Biological Activity and ADME Properties of Compounds
3.5. Softwares, Suites, and Webservers
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Newman, D.J.; Cragg, G.M.; Snader, K.M. Natural products as sources of new drugs over the period 1981-2002. J. Nat. Prod. 2003, 66, 1022–1037. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.H. Health benefits of fruit and vegetables are from additive and synergistic combinations of phytochemicals. Amer. J. Clin. Nutr. 2003, 78, 517–520. [Google Scholar] [CrossRef] [PubMed]
- Surh, Y.H. Cancer chemoprevention with dietary phytochemicals. Nat. Rev. Cancer 2003, 3, 768–780. [Google Scholar] [CrossRef] [PubMed]
- De Kok, T.M.; van Breda, S.G.; Manson, M.M. Mechanisms of combined action of different chemo preventive dietary compounds: A review. Eur. J. Nutr. 2008, 47, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Park, S. Cyclic glucans enhance solubility of bioavailable flavonoids. Molecules 2016, 21, 1556. [Google Scholar] [CrossRef] [PubMed]
- Sobeh, M.; Petruk, G.; Osman, S.; El Raey, M.A.; Imbimbo, P.; Monti, D.M.; Wink, M. Isolation of myricitrin and 3,5-di-O-mehtyl gossypetin from syzygiumsamarangense and evaluation of their involvement in protecting keratinocytes against oxidative stress via activation of the Nrf-2 pathway. Molecules 2019, 24, 1839. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhang, T. Antimicrobial activities of tea polyphenol on phytopathogens: A review. Molecules 2019, 24, 816. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.K.; Tsai, S.H.; Lin, S.Y. Anti-inflammatory and antitumor effects of flavonoids and flavanoids. Drugs Future 2001, 26, 145–152. [Google Scholar] [CrossRef]
- Mierziak, J.; Kostyn, K.; Kulma, A. Flavonoids as important molecules of plant interactions with the environment. Molecules 2014, 19, 16240–16265. [Google Scholar] [CrossRef]
- Mahomoodally, M.F.; Fakim, A.G.; Subratty, A.H. Antimicrobial activities and phytochemical profiles of endemic medicinal plants of Mauritius. Pharm. Biol. 2005, 43, 237–242. [Google Scholar] [CrossRef]
- Pandey, A.K. Anti-staphylococcal activity of a pan-tropical aggressive and obnoxious weed parthenium histerophorus: An in vitro study. Natl. Acad. Sci. Lett. 2007, 30, 383–386. [Google Scholar]
- Bohm, B.A. Introduction to Flavonoids; Gordon & Breach: Amsterdam, The Netherlands, 1998. [Google Scholar]
- Harborne, J.B.; Baxter, H. (Eds.) The Handbook of Natural Flavonoids; Wiley: Chichester, UK, 1999; Volume 1&2. [Google Scholar]
- Havsteen, B.H. The biochemistry and medical significance of the flavonoids. Pharm. Ther. 2002, 96, 67–202. [Google Scholar] [CrossRef]
- Skibola, C.F.; Smith, M.T. Potential health impacts of excessive flavonoid intake. Free Rad. Biol. Med. 2000, 29, 375–383. [Google Scholar] [CrossRef]
- Cazalolli, L.H.; Zanatta, L.; Alberton, E.H.; Figueiredo, M.S.R.B.; Folador, P.; Damazio, D.G.; Pizzolatti, F.; Silva, F.R.M.B. Flavonoids: Prospective drug candidates. Mini. Rev. Med. Chem. 2008, 8, 1429–1440. [Google Scholar] [CrossRef]
- Lalou, C.; Basak, P.; Mohantra, B.C.; Banik, R.; Dinda, B.; Khatib, A.M. Inhibition of tumor cells by the flavonoid furin inhibitor isolated from oroxylumindicum. Curr. Med. Chem. 2013, 20, 583–591. [Google Scholar] [PubMed]
- Suresh Babu, K.; HariBabu, T.; Srinivas, P.V.; Sastry, B.S.; Hara Kishore, K.; Murthy, U.S.N.; Rao, J.M. Synthesis and in vitro study of novel 7-O-acyl derivatives of Oroxylin A as antibacterial agents. Bioorg. Med. Chem. Lett. 2005, 15, 3953–3956. [Google Scholar] [CrossRef] [PubMed]
- Prakash, O.; Kumar, R.; Prakash, V. Synthesis and antifungal activity of some new 3-hydroxy-2-(1-phenyl-3-aryl-4-pyarazolyl) chromones. Eur. J. Med. Chem. 2008, 43, 435–440. [Google Scholar] [CrossRef] [PubMed]
- Losgen, S.; Magull, J.; Schulz, B.; Draeger, S.; Zeeck, A. Isofusidienols: Novel chromone-3-oxepines produced by the endophytic fungus Chalara sp. Eur. J. Org. Chem. 2008, 4, 698–703. [Google Scholar] [CrossRef]
- Ali, E.S.T.; Abdel-Aziz, S.A.G.; El-Shaaer, H.M.; Hanafy, F.I.; El-Fauomy, A.Z. Synthesis of some new 4-oxo-4H-chromene derivatives bearing nitrogen heterocyclic systems as antifungal agents. Turk. J. Chem. 2008, 32, 365–374. [Google Scholar]
- Hutter, J.A.; Salman, M.; Stavinoha, W.B.; Satangi, N.; Williams, R.F.; Streeper, R.T.; Weintraub, S.T. Anti-inflammatory C-glucosyl chromone from aloe barbadensis. J. Nat. Prod. 1996, 59, 541–543. [Google Scholar] [CrossRef]
- Mathias, L.; Silva, D.; Bernadete, P.; Mors, B.W.; Parente, J.P. Isolation and structural elucidation of a novel rotenoid from the seeds of clitoriafairchildiana. Nat. Prod. Res. 2005, 19, 325–329. [Google Scholar] [CrossRef] [PubMed]
- Birt, D.F.; Hendrich, S.; Wang, W. Dietary agents in cancer prevention: Flavonoids and isoflavonoids. Pharmacol. Ther. 2001, 90, 157–177. [Google Scholar] [CrossRef]
- Huang, W.; Ding, Y.; Miao, Y.; Liu, M.Z.; Li, Y.; Yang, G.F. Synthesis and antitumor activity of novel dithiocarbamate substituted chromones. Eur. J. Med. Chem. 2009, 44, 3687–3696. [Google Scholar] [CrossRef]
- Shaw, A.Y.; Chang, C.Y.; Liau, H.H.; Lu, P.J.; Chen, H.L.; Yang, C.N.; Li, H.Y. Synthesis of 2-styrylchromones as a novel class of antiproliferative agents targeting carcinoma cells. Eur. J. Med. Chem. 2009, 44, 2552–2562. [Google Scholar] [CrossRef] [PubMed]
- Pisco, L.; Kordian, M.; Peseke, K.; Feist, H.; Michalik, D.; Estrada, E.; Carvalho, J.; Hamilton, G.; Rando, D.; Quincoces, J. Synthesis of compounds with antiproliferative activity as analogues of prenylated natural products existing in Brazilian propolis. Eur. J. Med. Chem. 2006, 41, 401–407. [Google Scholar] [CrossRef] [PubMed]
- Kuroda, M.; Uchida, S.; Watanabe, K.; Mimaki, Y. Chromones form the tubers of eranthiscilicica and their antioxidant activity. Phytochemistry 2009, 70, 288–293. [Google Scholar] [CrossRef]
- Ly, C.; Yockell, L.J.; Ferraro, Z.M.; Arnason, J.T.; Ferrier, J.; Gruslin, A. The effects of dietary polyphenols on reproductive health and early development. Hum. Reprod. Update 2015, 21, 228–248. [Google Scholar] [CrossRef] [PubMed]
- Guo, K.; Liang, Z.; Liu, L.; Li, F.; Wang, H. Flavonoids intake and risk of prostate cancer: A meta-analysis of observational studies. Andrologia 2016, 48, 1175–1182. [Google Scholar] [CrossRef] [PubMed]
- Qais, N.; Rahman, M.M.; Rashid, M.A.; Koshino, H.; Nagasawa, K.; Nakata, T. A new C-benzylated chalcone from Desmos chinensis. Fitoterpia 1996, 67, 511–514. [Google Scholar]
- Fishkin, R.J.; Winslow, J.T. Endotoxin-induced reduction of social investigation by mice: Interaction with amphetamine and anti-inflammatory drugs. Psychopharmacology (Berl.) 1997, 132, 335–341. [Google Scholar] [CrossRef]
- Pearce, F.L.; Befus, A.D.; Bienenstock, J. Mucosal mast cells: III. Effect of quercetin and other flavonoids on antigen-induced histamine secretion from rat intestinal mast cells. J. Allergy. Clin. Immunol. 1984, 73, 819–823. [Google Scholar] [CrossRef]
- Hecker, M.; Preiss, C.; Klemm, P.; Busse, R. Inhibition by antioxidants of nitric oxide synthase expression in murine macrophages: Role of nuclear factor kB and interferon regulatory factor 1”. Br. J. Pharmcol. 1996, 118, 2178–2184. [Google Scholar] [CrossRef]
- Cardenas, M.; Marder, M.; Blank, V.C.; Roguin, L.P. Antitumor activity of some natural flavonoids and synthetic derivatives on various human and murine cancer cell lines. Bioorg. Med. Chem. 2006, 14, 2966–2971. [Google Scholar] [CrossRef] [PubMed]
- Wei, F.; Jubo, W.; Liqin, Y.; Li, Z.; Na, L.; Qidong, Y.; Qinglong, G.; Zhiyu, L. Synthesis and biological evaluation of 7-O-modified oroxylin A derivatives. Bioorg. Med. Chem. Lett. 2012, 22, 1118–1121. [Google Scholar]
- Kun, H.; Wei, W.; Hong, C.; Sha, S.P.; Jie, R. Synthesis and cytotoxicity of novel chrysin derivatives. Med. Chem. Res. 2011, 20, 838–846. [Google Scholar]
- Li, H.Q.; Shi, L.; Li, Q.S.; Liu, P.G.; Luo, Y.; Zhao, J.; Zhu, H.L. Synthesis of C(7) modified chrysin derivatives designing to inhibit β-ketoacyl carrier protein synthase III (FabH) as antibiotics. Bioorg. Med. Chem. 2009, 17, 6264–6269. [Google Scholar] [CrossRef] [PubMed]
- Bonini, C.; Righi, G. Regio- and chemoselective synthesis of halohydrins by cleavage of oxiranes with metal hydrides. Synthesis 1994, 3, 225–238. [Google Scholar] [CrossRef]
- Azizi, N.; Pourhasan, B.; Aryanasab, F.; Saidi, M.R. A simple and novel eco-friendly process for the one-pot synthesis of dithiocarbamates from amines, carbon disulphide and epoxides. Synlett 2007, 8, 1239–1242. [Google Scholar]
- Azizi, N.; Gholibeglo, E.; Maryami, M.; Nayeri, S.D.; Bolourtchian, S.M. Ultrasound mediated efficient ring opening of epoxides by in situ generated dithiocarbamates in green reaction media. C. R. Chim. 2013, 16, 412–418. [Google Scholar] [CrossRef]
- Morf, P.; Raimondi, F.; Nothofer, H.G.; Schnyder, B.; Yasuda, A.; Wessels, J.M.; Jung, T.A. Dithiocarbamates: Functional and versatile linkers for the formation of self-assembled monolayers. Langmuir 2006, 22, 658–663. [Google Scholar] [CrossRef]
- McClain, A.; Hsieh, Y.L. Synthesis of polystyrene-supported dithiocarbamates and their complexation with metal ions. J. Appl. Polym. Sci. 2004, 92, 218–225. [Google Scholar] [CrossRef]
- Dunn, A.D.; Rudorf, W.D. Carbon Disulphide in Organic Chemistry; Ellis Hordwood: Chichester, UK, 1989; pp. 226–367. [Google Scholar]
- Griffin, T.S.; Woods, T.S.; Klayman, D.L. Thioureas in the synthesis of Heterocycles. In Advances in Heterocyclic Chemistry; Academic Press Inc., Elsevier: Amsterdam, The Netherlands, 1975; Volume 18, pp. 99–158. [Google Scholar]
- Liao, S.; Raung, S.; Chen, C. Japanese encephalitis virus stimulates superoxide dismutase activity in rat glial cultures. Neurosci. Lett. 2002, 324, 133–136. [Google Scholar] [CrossRef]
- Gursoy, A.; Ates, O.; Karali, N.; Cesur, N.; Kiraz, M. Synthesis and antifungal activity of new carbamodithioc acid esters derived from 3-acetylcoumarin. Eur. J. Med. Chem. 1996, 31, 643–646. [Google Scholar] [CrossRef]
- Ronconi, L.; Marzano, C.; Zanello, P.; Corsini, M.; Miolo, G.; Macca, C.; Trevisan, A.; Fregona, D. Gold (III) dithiocarbamate derivatives for the treatment of cancer: Solution chemistry, DNA binding and hemolytic properties. J. Med. Chem. 2006, 49, 1648–1657. [Google Scholar] [CrossRef] [PubMed]
- Bandari, S.K.; Kammari, B.R.; Madda, J.; Kommu, N.; Lakkadi, A.; Vuppala, S.; Tigulla, P. Synthesis of new chromeno-carbamodithioate derivatives and preliminary evaluation of their antioxidant activity and molecular docking studies. Bioorg. Med. Chem. Lett. 2017, 27, 1256–1260. [Google Scholar] [CrossRef]
- Thorn, G.D.; Ludwig, R.A. The Dithiocarbamates and Related Compounds; Elsvier: Amsterdam, The Netherlands, 1962. [Google Scholar]
- Nace, H.R. The preparation of olefins by the pyrolysis of xanthates: The chugaev reaction. Org. React. 1962, 12, 57. [Google Scholar]
- Rafin, C.; Veignie, E.; Sanchole, M.; Postel, D.; Len, C.; Villa, P.; Ronco, G. Synthesis and antifungal activity of novel bisdithiocarbamate derivatives of carbohydrates against fusarium oxysporum f. sp. lini. J Agric. Food Chem. 2000, 48, 5283–5287. [Google Scholar] [CrossRef]
- Ramesh, P.; Sanjeeva Reddy, C.; Suresh Babu, K.; Muralidhar Reddy, P.; SrinivasaRao, V.; Parthasarathy, T. Synthesis, characterization and molecular docking studies of novel 2-amino 3-cyano pyrano [2,3H]chrysin derivatives as potential antimicrobial agents. Med. Chem. Res. 2015, 24, 3696–3709. [Google Scholar] [CrossRef]
- Suresh Babu, K.; HariBabu, T.; Srinivas, P.V.; Hara Kishore, K.; Rao, J.M. Synthesis and biological evaluation of novel C (7) modified chrysin analogues as antibacterial agents. Bioorg. Med. Chem. Lett. 2006, 16, 221–224. [Google Scholar] [CrossRef]
- Patel, M.A.; Bhila, V.G.; Patel, N.H.; Patel, A.K.; Brahmbhatt, D.I. Synthesis, characterization and biological evaluation of some pyridine and quinoline fused chromenone derivatives. Med. Chem. Res. 2012, 21, 4381–4388. [Google Scholar] [CrossRef]
- Shridhar, R.; Perumal, P.T.; Etti, S.; Shanmugam, G.; Ponnuswamy, M.N.; Prabavathy, V.R.; Mathivanan, N. Design, synthesis and anti-microbial activity of 1H-pyrazole carboxylates. Bioorg. Med. Chem. Lett. 2004, 14, 6035–6040. [Google Scholar] [CrossRef]
- Keche, A.P.; Hatnapure, G.D.; Tale, R.T.; Rodge, A.H.; Birajdar, S.S.; Kamble, V.M. Synthesis, anti-inflammatory and antimicrobial evaluation of novel N1-(quinolin-4yl)ethane-1,2-diamine phenyl urea derivatives. Med. Chem. Res. 2013, 22, 1480–1487. [Google Scholar] [CrossRef]
- Shamroukh, A.H.; Zaki, M.E.A.; Morsy, E.M.H.; Abdel Motti, F.M.; Abdel Megeid, F.M.E. Synthesis, isomerization and antimicrobial evaluation of some pyrazolopyranotriazolopyrimindine derivatives. Arch. Pharm. 2007, 340, 345–351. [Google Scholar] [CrossRef]
- Khandekar, S.S.; Daines, R.A.; Lonsdale, J.T. Bacterial β-Ketoacyl-acyl carrier protein synthases as targets for antibacterial agents. Curr. Protein Pept. Sci. 2003, 4, 21–29. [Google Scholar] [CrossRef]
- Heath, R.J.; Rock, C.O. The claisen condensation in biology. Nat. Prod. Rep. 2002, 19, 581–596. [Google Scholar] [CrossRef]
- Tsay, J.T.; Oh, W.; Larson, T.J.; Jakowski, S.; Rock, C.O. Isolation and characterization of the beta-ketoacyl-acyl carrier protein synthase III gene (fabH) form Escherichia coli K-12. J. Biol. Chem. 1992, 267, 6807–6814. [Google Scholar]
- Clough, R.C.; Matthis, A.L.; Barnum, S.R.; Jaworski, J.G. Purification and characterization of 3-ketoacyl-acyl carrier protein synthase III from spinach. A condensing enzyme utilizing acetyl-coenzyme A to initiate fatty acid synthesis. J. Biol. Chem. 1992, 267, 20992–20998. [Google Scholar]
- Heath, R.J.; Rock, C.O. Regulation of fatty acid elongation and initiation by acyl-acyl carrier protein in Escherichia coli. J. Biol. Chem. 1996, 271, 1833–1836. [Google Scholar] [CrossRef]
- Molinspiration Cheminformatics, Bratislava, Slovak Republic. Available online: http://www.molinspiration.com/services/properties.html (accessed on 22 April 2010).
- Lipinski, C.A.; Lombardo, L.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimated solubility and permeability in drug discovery and development settings. Adv. Drug Deliv.Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Zhao, Y.H.; Abraham, M.H.; Le, J. Rate-limited steps of human oral absorption and QSAR studies. Pharm. Res. 2002, 19, 1446–1457. [Google Scholar] [CrossRef]
- Drug-Likeness and Molecular Property Prediction. Available online: http://www.molsoft.com/mptop (accessed on 11 April 2019).
- Devi, A.M.; Gopinath, G.; Srinivas, B.; Venu, S.; Madhavi, M.; Sudha, N.S.; Sudhakar, K.; Rani, A.R.; Rao, S.S. Virtual screening of RAGE inhibitors using molecular docking. Bioinformation 2016, 12, 124–130. [Google Scholar]
- Jorgensen, W.L.; Tirado-Rives, J. Potential energy functions for atomic level simulations of water and organic and bimolecular systems. Proc. Nat. Acad. Sci. USA 2005, 102, 6665–6670. [Google Scholar] [CrossRef]
- LigPrep; Version 2.3; Schrodinger, L.L.C.: New York, NY, USA, 2009.
- Brown, A.; Long, F.; Nicholis, R.A.; Toots, J.; Emsley, P.; Murshudov, G. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr. D Biol. Crystallogr. 2015, 1, 136–153. [Google Scholar] [CrossRef]
- Brooks, R.R.; Brooks, C.L.; Mackerell, A.D., Jr.; Nilsson, L.; Petrella, R.J.; Roux, B.; Won, Y.; Archontis, G.; Bartels, C.; Boresch, S.; et al. CHARMM: The biomolecular simulation program. J. Comp. Chem. 2009, 30, 1545–1614. [Google Scholar]
- Thomsen, R.; Christensen, M.H. MolDock: A new technique for high-accuracy molecular docking. J. Med. Chem. 2006, 49, 3315–3321. [Google Scholar] [CrossRef]
- Nelder, J.A.; Mead, R. A simplex method for function minimization. Comput. J. 1965, 7, 308–313. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds 3a, 3c, 3d, 3f and 3h are available from the authors. |
| S.No | Entry | HNR1R2 | Product b | Yield c (%) | M.P (°C) |
|---|---|---|---|---|---|
| 1 | 3a | Piperazine | 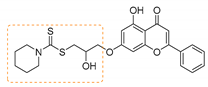 | 89 | 134–136 |
| 2 | 3b | Pyrrolidine | 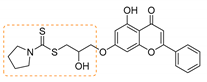 | 88 | 144–146 |
| 3 | 3c | Morpholine |  | 88 | 146–148 |
| 4 | 3d | 4-Benzyl piperazine |  | 81 | 154–156 |
| 5 | 3e | Thiomorpholine | 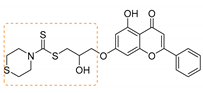 | 82 | 168–170 |
| 6 | 3f | 4-Fluorophenyl piperazine | 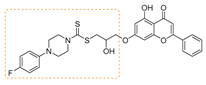 | 74 | 146–148 |
| 7 | 3g | 4-Pyridyl piperazine | 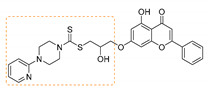 | 68 | 152–154 |
| 8 | 3h | 4-Methoxyphenyl piperazine | 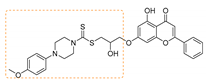 | 73 | 151–152 |
| 9 | 3i | Cis-3,5-Dimethyl morpholine |  | 71 | 181–183 |
| 10 | 3j | 4-Benzyloxy carbonyl piperazine |  | 66 | 140–142 |
| 11 | 3k | Diethyl amine | 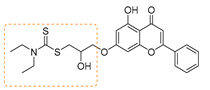 | 85 | 158–160 |
| Zone of Inhibition a (mm) and MIC b (μg/mL) Values of Compounds | ||||||
|---|---|---|---|---|---|---|
| Gram-Positive Bacteria | Gram-Negative Bacteria | Fungi | ||||
| Compound Code | S. epidermidis | B. subtilis | E. coli | P. aeruginosa | S. cerevisiae | C. albicans |
| 3a | 16 | 14 | 17 | 10 | 8 | 6 |
| 3b | 14 | 10 | 11 | 13 | 7 | 8 |
| 3c | 10 | 8 | 9 | 11 | 9 | 6 |
| 3d | 26 (9.37) | 30 (4.68) | 29 (4.68) | 23 (18.75) | 23 (18.75) | 21 (18.75) |
| 3e | 18 | 19 | 22 | 19 | 8 | 9 |
| 3f | 23 (18.75) | 26 (9.37) | 25 (9.37) | 21 (18.75) | 27 (9.37) | 21 (18.75) |
| 3g | 21 (18.75) | 18 (18.75) | 19 (9.37) | 20 (18.75) | 9 | 7 |
| 3h | 21 | 17 | 20 | 19 | 28 (4.68) | 26 (18.75) |
| 3i | 9 | 7 | 10 | 7 | 5 | 8 |
| 3j | 24 (9.37) | 23 (4.68) | 21 (9.37) | 20 (18.75) | 21 (18.75) | 19 (37.5) |
| 3k | 7 | 9 | 12 | 10 | 13 (18.75) | 14 (37.5) |
| 1 | 6 | 5 | 8 | 6 | - | - |
| 2 | 7 | 8 | 9 | 7 | - | - |
| Penicillin | 33 (2.34) | 35 (1.17) | 29 (9.37) | 28 (9.37) | - | - |
| Itrazole | - | - | - | - | 31 (1.17) | 28 (9.37) |
| S. No | Ligand | Moldock Score [Grid](kcal/mol) | Moldock Score | Rerank Score | RMSD |
|---|---|---|---|---|---|
| 1 | Penicillin | −140.42 | −141.09 | −97.99 | 51.88 |
| 2 | Chrysin | −95.05 | −97.72 | −24.61 | 44.38 |
| 3 | 2 | −100.46 | −97.65 | −76.56 | 53.37 |
| 4 | 3a | −137.01 | −140.03 | −125.10 | 53.53 |
| 5 | 3b | −139.01 | −139.60 | −100.94 | 55.87 |
| 6 | 3c | −135.46 | −136.85 | −118.25 | 53.98 |
| 7 | 3d | −156.02 | −158.49 | −100.42 | 49.82 |
| 8 | 3e | −146.08 | −146.70 | −130.27 | 53.84 |
| 9 | 3f | −152.07 | −153.97 | −130.76 | 59.85 |
| 10 | 3g | −151.49 | −155.46 | −132.91 | 51.90 |
| 11 | 3h | −148.68 | −146.88 | −110.41 | 50.82 |
| 12 | 3i | −128.11 | −133.98 | −47.94 | 50.36 |
| 13 | 3j | −154.52 | −157.49 | −102.96 | 43.40 |
| 14 | 3k | −137.98 | −139.31 | −108.02 | 53.31 |
| S. No | Ligand | Moldock Score [Grid](kcal/mol) | Moldock Score | Rerank Score | RMSD |
|---|---|---|---|---|---|
| 1 | Itrazole | −213.68 | −218.38 | −171.75 | 38.25 |
| 2 | Chrysin | −96.56 | −94.56 | −81.26 | 38.67 |
| 3 | 2 | −116.34 | −115.26 | −97.75 | 32.62 |
| 4 | 3a | −157.20 | −154.09 | −127.09 | 35.42 |
| 5 | 3b | −169.85 | −170.41 | −140.99 | 35.93 |
| 6 | 3c | −161.94 | −164.89 | −134.37 | 29.90 |
| 7 | 3d | −184.72 | −186.61 | −154.64 | 29.64 |
| 8 | 3e | −167.83 | −170.79 | −143.70 | 30.06 |
| 9 | 3f | −186.76 | −189.74 | −155.25 | 36.50 |
| 10 | 3g | −176.23 | −173.46 | −128.58 | 38.73 |
| 11 | 3h | −190.54 | −192.50 | −157.04 | 29.60 |
| 12 | 3i | −160.31 | −161.26 | −116.69 | 36.54 |
| 13 | 3j | −187.92 | −189.65 | −141.46 | 28.25 |
| 14 | 3k | −165.14 | −164.57 | −140.99 | 36.04 |
| Comp | Gpcr Ligand | Ion Channel Modulator | Kinase Inhibitor | Nuclear Receptor Ligand | Protease Inhibitor | Enzyme Inhibitor | milogP [a] | TPSA (A2) [b] | n Violation [c] | M.wt [d] | nON [e] | nOHNH [f] | %ABS | MV [g] |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Rule | ≤5 | --- | ≤1 | <500 | <10 | <5 | ||||||||
| Penicillin | 0.01 | −0.42 | −0.71 | −0.37 | 0.86 | 0.30 | 1.82 | 86.71 | 0 | 334.40 | 6 | 2 | 79.08 | 287.55 |
| Chrysin | −0.11 | −0.08 | 0.15 | 0.30 | −0.30 | 0.26 | 2.94 | 70.67 | 0 | 254.24 | 4 | 2 | 84.61 | 216.03 |
| 2 | −0.03 | −0.35 | 0.26 | 0.21 | 0.03 | 0.21 | 3.31 | 72.20 | 0 | 310.31 | 5 | 1 | 84.09 | 265.57 |
| 3a | −0.12 | −0.54 | −0.31 | −0.21 | −0.28 | 0.07 | 4.70 | 83.14 | 0 | 471.60 | 6 | 2 | 80.31 | 407.39 |
| 3b | −0.12 | −0.54 | −0.29 | −0.19 | −0.26 | 0.01 | 4.20 | 83.14 | 0 | 457.57 | 6 | 2 | 80.31 | 390.59 |
| 3c | −0.18 | −0.61 | −0.29 | −0.24 | −0.31 | 0.04 | 3.64 | 92.37 | 0 | 473.57 | 7 | 2 | 77.13 | 399.57 |
| 3d | −0.11 | −0.62 | −0.27 | −0.25 | −0.26 | 0.01 | 5.08 | 86.38 | 2 | 562.71 | 7 | 2 | 79.19 | 491.58 |
| 3e | −0.14 | −0.55 | −0.29 | −0.24 | −0.28 | 0.08 | 4.18 | 83.14 | 0 | 489.64 | 6 | 2 | 80.31 | 408.71 |
| 3f | −0.10 | −0.69 | −0.28 | −0.27 | −0.27 | −0.04 | 5.25 | 86.38 | 2 | 580.70 | 7 | 2 | 79.18 | 496.51 |
| 3g | −0.04 | −0.51 | −0.13 | −0.27 | −0.26 | 0.09 | 4.48 | 99.27 | 1 | 549.67 | 8 | 2 | 74.75 | 470.62 |
| 3h | −0.14 | −0.71 | −0.30 | −0.30 | −0.32 | −0.09 | 5.44 | 95.61 | 2 | 578.71 | 8 | 2 | 76.01 | 500.32 |
| 3i | −0.09 | −0.48 | −0.23 | −0.13 | −0.20 | 0.02 | 4.30 | 92.37 | 1 | 501.63 | 7 | 2 | 77.13 | 432.75 |
| 3j | −0.03 | −0.50 | −0.27 | −0.12 | −0.06 | 0.09 | 4.72 | 112.68 | 1 | 572.71 | 9 | 2 | 70.12 | 497.52 |
| 3k | −0.17 | −0.60 | −0.34 | −0.24 | −0.35 | 0.04 | 4.55 | 83.14 | 0 | 459.59 | 6 | 2 | 80.31 | 400.95 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramesh, P.; Srinivasa Rao, V.; Hong, Y.-A.; Reddy, P.M.; Hu, A. Molecular Design, Synthesis, and Biological Evaluation of 2-Hydroxy-3-Chrysino Dithiocarbamate Derivatives. Molecules 2019, 24, 3038. https://doi.org/10.3390/molecules24173038
Ramesh P, Srinivasa Rao V, Hong Y-A, Reddy PM, Hu A. Molecular Design, Synthesis, and Biological Evaluation of 2-Hydroxy-3-Chrysino Dithiocarbamate Derivatives. Molecules. 2019; 24(17):3038. https://doi.org/10.3390/molecules24173038
Chicago/Turabian StyleRamesh, Pulabala, Vankadari Srinivasa Rao, Yi-An Hong, P. Muralidhar Reddy, and Anren Hu. 2019. "Molecular Design, Synthesis, and Biological Evaluation of 2-Hydroxy-3-Chrysino Dithiocarbamate Derivatives" Molecules 24, no. 17: 3038. https://doi.org/10.3390/molecules24173038
APA StyleRamesh, P., Srinivasa Rao, V., Hong, Y.-A., Reddy, P. M., & Hu, A. (2019). Molecular Design, Synthesis, and Biological Evaluation of 2-Hydroxy-3-Chrysino Dithiocarbamate Derivatives. Molecules, 24(17), 3038. https://doi.org/10.3390/molecules24173038