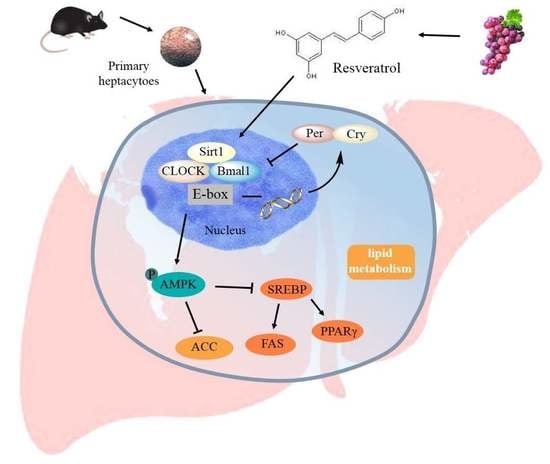
Resveratrol Maintains Lipid Metabolism Homeostasis via One of the Mechanisms Associated with the Key Circadian Regulator Bmal1
Abstract
1. Introduction
2. Results
2.1. Intervention Effect of RES on Morphology Changes and Intracellular Lipid Accumulation under High Fatty Acid Conditions
2.2. RES Ameliorates FFA-Triggered Hepatic Glucose Metabolism and Lipid Metabolism Unbalance
2.3. RES Attenuates FFA-Triggered Oxidative Stress and Mitochondrial Dysfunction in Hepatic HepG2 Cells.
2.4. RES-Modulated Circadian Misalignment in Metabolic Disorders
2.5. RES Eliminated FFA-Induced Lipid Metabolism Imbalance by Regulating the Circadian Clock in Hepatic HepG2 Cells
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Treatment
4.2. Isolation and Treatment of Primary Murine Hepatocytes
4.3. Small Interfering RNA (siRNA) Transfection of Hepatocytes
4.4. Synchronization of HepG2 Cells
4.5. Cell Viability
4.6. Lipid Levels of Hepatocytes
4.7. Mitochondrial Membrane Potential (MMP) Assay
4.8. Detection of Reactive Oxygen Species (ROS)
4.9. RNA Preparation and Quality Control and Real-Time Quantitative PCR (RT-qPCR)
4.10. Western Blot
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
References
- Armitage, J.A.; Poston, L.; Taylor, P.D. Developmental origins of obesity and the metabolic syndrome: The role of maternal obesity. Front. Horm. Res. 2008, 36, 73–84. [Google Scholar] [PubMed]
- Gokhan, S. Hotamisligil: Inflammation and metabolic disorders. Nature 2006, 444, 860–867. [Google Scholar]
- Feldstein, A.E.; Werneburg, N.W.; Canbay, A.; Guicciardi, M.E.; Bronk, S.F.; Rydzewski, R.; Burgart, L.J.; Gores, G.J. Free fatty acids promote hepatic lipotoxicity by stimulating TNF-alpha expression via a lysosomal pathway. Hepatology 2004, 40, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Donnelly, K.L.; Smith, C.I.; Schwarzenberg, S.J.; Jessurun, J.; Boldt, M.D.; Parks, E.J. Sources of fatty acids stored in liver and secreted via lipoproteins in patients with nonalcoholic fatty liver disease. J. Clin. Invesig. 2005, 115, 1343–1351. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Barnes, G.T.; Yang, Q.; Tan, G.; Yang, D.; Chou, C.J.; Sole, J.; Nichols, A.; Ross, J.S.; Tartaglia, L.A.; et al. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J. Clin. Invesig. 2003, 112, 1821–1830. [Google Scholar] [CrossRef]
- Hirosumi, J.; Tuncman, G.; Chang, L.; Görgün, C.Z.; Uysal, K.T.; Maeda, K.; Karin, M.; Hotamisligil, G.S. A central role for JNK in obesity and insulin resistance. Nature 2002, 420, 333–336. [Google Scholar] [CrossRef]
- Utzschneider, K.M.; Kahn, S.E. Review: The role of insulin resistance in nonalcoholic fatty liver disease. J. Clin. Endocr. Metab. 2006, 91, 4753–4761. [Google Scholar] [CrossRef]
- Rutter, J.; And, M.R.; McKnight, S.L. Metabolism and the Control of Circadian Rhythms. Annu. Rev. Biochem. 2002, 71, 307–331. [Google Scholar] [CrossRef]
- Kristin, E.; Paolo, S. Metabolism and the circadian clock converge. Physiol. Rev. 2013, 93, 107–135. [Google Scholar]
- Cho, H.; Zhao, X.; Hatori, M.; Yu, R.T.; Barish, G.D.; Lam, M.T.; Chong, L.W.; DiTacchio, L.; Atkins, A.R.; Glass, C.K.; et al. Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature 2012, 485, 123–127. [Google Scholar] [CrossRef]
- Shearman, L.P.; Sriram, S.; Weaver, D.R.; Maywood, E.S.; Chaves, I.; Zheng, B.; Kume, K.; Lee, C.C.; Gt, V.D.H.; Hastings, M.H. Interacting molecular loops in the mammalian circadian clock. Science 2000, 288, 1013–1019. [Google Scholar] [CrossRef]
- Tan, X.; Zhao, T.; Wang, Z.; Wang, J.; Wang, Y.; Liu, Z.; Liu, X. Acrylamide Defects the Expression Pattern of the Circadian Clock and Mitochondrial Dynamics in C57BL/6J Mice Liver and HepG2 Cells. J. Agric. Food Chem. 2018, 66, 10252–10266. [Google Scholar] [CrossRef]
- Tan, X.; Ye, J.; Liu, W.; Zhao, B.; Shi, X.; Zhang, C.; Liu, Z.; Liu, X. Acrylamide aggravates cognitive deficits at night period via the gut–brain axis by reprogramming the brain circadian clock. Arch. Toxicol. 2018, 1–20. [Google Scholar] [CrossRef]
- Orozcosolis, R.; Aguilararnal, L.; Murakami, M.; Peruquetti, R.; Ramadori, G.; Coppari, R.; Sassonecorsi, P. The Circadian Clock in the Ventromedial Hypothalamus Controls Cyclic Energy Expenditure. Cell Metab. 2016, 23, 467–478. [Google Scholar] [CrossRef]
- Pan, X.; Zhang, Y.; Wang, L.; Hussain, M.M. Diurnal regulation of MTP and plasma triglyceride by CLOCK is mediated by SHP. Cell Metab. 2010, 12, 174–186. [Google Scholar] [CrossRef]
- Tseng, H.L.; Yang, S.C.; Yang, S.H.; Shieh, K.R. Hepatic circadian-clock system altered by insulin resistance, diabetes and insulin sensitizer in mice. PLoS ONE 2015, 10, e0120380. [Google Scholar] [CrossRef][Green Version]
- Kristin, E.; Paolo, S. Metabolism control by the circadian clock and vice versa. Nat. Struct. Mol. Biol. 2009, 16, 462–467. [Google Scholar]
- Turek, F.W.; Joshu, C.; Kohsaka, A.; Lin, E.; Ivanova, G.; McDearmon, E.; Eckel, R.H.; Laposky, A.; Losee-Olson, S.; Easton, A. Obesity and metabolic syndrome in circadian Clock mutant mice. Science 2005, 308, 1043–1045. [Google Scholar] [CrossRef]
- Green, C.B.; Takahashi, J.S.; Bass, J. The Meter of Metabolism. Cell 2008, 134, 728–742. [Google Scholar] [CrossRef]
- Kohsaka, A.; Laposky, A.D.; Ramsey, K.M.; Estrada, C.; Joshu, C.; Kobayashi, Y.; Turek, F.W.; Bass, J. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007, 6, 414–421. [Google Scholar] [CrossRef]
- Guo, R.; Zhao, B.; Wang, Y.; Wu, D.; Wang, Y.; Yu, Y.; Yan, Y.; Zhang, W.; Liu, Z.; Liu, X. Cichoric acid prevents free fatty acids induced lipid metabolism disorders via regulating bmal1 in HepG2 cells. J. Agric. Food Chem. 2018, 66, 9667–9678. [Google Scholar] [CrossRef]
- Qi, G.; Guo, R.; Tian, H.; Li, L.; Liu, H.; Mi, Y.; Liu, X. Nobiletin protects against insulin resistance and disorders of lipid metabolism by reprogramming of circadian clock in hepatocytes. BBA-Mol. Cell Biol. Lipids 2018, 1863, 549–562. [Google Scholar] [CrossRef]
- Mi, Y.; Qi, G.; Gao, Y.; Li, R.; Wang, Y.; Li, X.; Huang, S.; Liu, X. EGCG Ameliorates Insulin Resistance and Mitochondrial Dysfunction in HepG2 Cells: Involvement of Bmal1. Mol. Nutr. Food Res. 2017, 61, 1700440. [Google Scholar]
- Jang, M.; Cai, L.; Udeani, G.O.; Slowing, K.V.; Thomas, C.F.; Beecher, C.W.; Fong, H.H.; Farnsworth, N.R.; Kinghorn, A.D.; Mehta, R.G. Cancer chemopreventive activity of resveratrol, a natural product derived from grapes. Science 1997, 275, 218–220. [Google Scholar] [CrossRef]
- Chen, S.; Zhao, X.; Ran, L.; Wan, J.; Wang, X.; Qin, Y.; Shu, F.; Gao, Y.; Yuan, L.; Zhang, Q. Resveratrol improves insulin resistance, glucose and lipid metabolism in patients with non-alcoholic fatty liver disease: A randomized controlled trial. Digest Liver Dis. 2015, 47, 226–232. [Google Scholar] [CrossRef]
- Sun, L.; Wang, Y.; Song, Y.; Cheng, X.R.; Xia, S.; Rahman, M.R.; Shi, Y.; Le, G. Resveratrol restores the circadian rhythmic disorder of lipid metabolism induced by high-fat diet in mice. Biochem Biophys Res Commun. 2015, 458, 86–91. [Google Scholar] [CrossRef]
- Estany, S.; Palacio, J.R.; Barnadas, R.; Sabes, M.; Iborra, A.; Martínez, P. Antioxidant activity of N-acetylcysteine, flavonoids and alpha-tocopherol on endometrial cells in culture. J. Reprod. Immunol. 2007, 75, 1–10. [Google Scholar] [CrossRef]
- Marcsek, Z.L.; Kocsis, Z.; Szende, B.; Tompa, A. Effect of formaldehyde and resveratrol on the viability of Vero, HepG2 and MCF-7 cells. Cell Biol Int. 2007, 31, 1214–1219. [Google Scholar] [CrossRef]
- Li, Y.; Zhu, W.; Li, J.; Liu, M.; Wei, M. Resveratrol suppresses the STAT3 signaling pathway and inhibits proliferation of high glucose-exposed HepG2 cells partly through SIRT1. Oncol. Rep. 2013, 30, 2820–2828. [Google Scholar] [CrossRef]
- Ou, X.; Chen, Y.; Cheng, X.; Zhang, X.; He, Q. Potentiation of resveratrol-induced apoptosis by matrine in human hepatoma HepG2 cells. Oncol. Rep. 2014, 32, 2803–2809. [Google Scholar] [CrossRef]
- Cretene, G.; Clech, M.L.; Gachon, F. Circadian clock-coordinated 12 Hr period rhythmic activation of the IRE1alpha pathway controls lipid metabolism in mouse liver. Cell Metab. 2010, 11, 47–57. [Google Scholar] [CrossRef]
- Eckel-Mahan, K.L.; Patel, V.R.; De, M.S.; Orozco-Solis, R.; Ceglia, N.J.; Sahar, S.; Dilag-Penilla, S.A.; Dyar, K.A.; Baldi, P.; Sassone-Corsi, P. Reprogramming of the circadian clock by nutritional challenge. Cell 2013, 155, 1464–1478. [Google Scholar] [CrossRef]
- Panda, S. Circadian physiology of metabolism. Science 2016, 354, 1008–1015. [Google Scholar] [CrossRef]
- Shi, S.; Ansari, T.S.; Mcguinness, O.P.; Wasserman, D.H.; Johnson, C.H. Circadian Disruption Leads to Insulin Resistance and Obesity. Curr. Biol. 2013, 23, 372–381. [Google Scholar] [CrossRef]
- Lemmer, B. The importance of circadian rhythms on drug response in hypertension and coronary heart disease--from mice and man. Pharmacol. Therapeut. 2006, 111, 629–651. [Google Scholar] [CrossRef]
- Msa, B.; Lee, D.H.; Kim, H.J.; Lee, S.H.; Cho, S.H.; Yang, B.S.; Kim, S.D. Estimates of genetic parameters for fatty acid compositions in the longissimus dorsi muscle of Hanwoo cattle. Animal 2017, 12, 675–683. [Google Scholar]
- He, B.; Nohara, K.; Park, N.; Park, Y.S.; Guillory, B.; Zhao, Z.; Garcia, J.M.; Koike, N.; Lee, C.C.; Takahashi, J.S. The small molecule nobiletin targets the molecular oscillator to enhance circadian rhythms and protect against metabolic syndrome. Cell Metab. 2016, 23, 610–621. [Google Scholar] [CrossRef]
- Antoch, M.P.; Kondratov, R.V. Pharmacological modulators of the circadian clock as potential therapeutic drugs: Focus on genotoxic/anticancer therapy. Springer 2013, 217, 289–309. [Google Scholar]
- Qi, G.; Mi, Y.; Liu, Z.; Fan, R.; Qiao, Q.; Sun, Y.; Ren, B.; Liu, X. Dietary tea polyphenols ameliorate metabolic syndrome and memory impairment via circadian clock related mechanisms. J. Funct. Foods 2017, 34, 168–180. [Google Scholar] [CrossRef]
- Froy, O. The relationship between nutrition and circadian rhythms in mammals. Front. Neuroendocrin 2007, 28, 61–71. [Google Scholar] [CrossRef]
- Bi, X.; Liu, W.; Xia, X.; Chen, L.; Mu, T.; Liu, J.; Hou, Y.; Zhao, Y. Effects of Callistephus chinensis flower polyphones on improving metabolic disorders in high-fat diet-induced mice. Food Funct. 2014, 5, 3304–3310. [Google Scholar] [CrossRef]
- Inzaugarat, M.E.; De, M.E.; Baz, P.; Lucero, D.; García, C.C.; Gonzalez, B.E.; Daruich, J.; Sorda, J.A.; Wald, M.R.; Cherñavsky, A.C. New evidence for the therapeutic potential of curcumin to treat nonalcoholic fatty liver disease in humans. PLoS ONE 2017, 12, e0172900. [Google Scholar] [CrossRef]
- Musso, G.; Gambino, R.; Cassader, M. Non-alcoholic fatty liver disease from pathogenesis to management: An update. Obes. Rev. 2010, 11, 430–445. [Google Scholar] [CrossRef]
- Malhi, H.; Bronk, S.F.; Werneburg, N.W.; Gores, G.J. Free fatty acids induce JNK-dependent hepatocyte lipoapoptosis. J. Biol. Chem. 2006, 281, 12093–12101. [Google Scholar] [CrossRef]
- Zhou, B.; Zhou, D.; Wei, X.; Zhong, R.; Xu, J.; Sun, L. Astragaloside IV attenuates free fatty acid-induced ER stress and lipid accumulation in hepatocytes via AMPK activation. Acta Pharmacol. Sin. 2017, 38, 998–1008. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhou, B.; Zhang, F.; Wu, J.; Hu, Y.; Liu, Y.; Zhai, Q. Amyloid-β induces hepatic insulin resistance by activating JAK2/STAT3/SOCS-1 signaling pathway. Diabetes 2012, 61, 1434–1443. [Google Scholar] [CrossRef]
- Zwighaft, Z.; Aviram, R.; Shalev, M.; Roussonoori, L.; Krautcohen, J.; Golik, M.; Brandis, A.; Reinke, H.; Aharoni, A.; Kahana, C. Circadian Clock Control by Polyamine Levels through a Mechanism that Declines with Age. Cell Metab. 2015, 22, 874–885. [Google Scholar] [CrossRef]
- Wang, Y.; Diao, Z.; Li, J.; Ren, B.; Zhu, D.; Liu, Q.; Liu, Z.; Liu, X. Chicoric acid supplementation ameliorates cognitive impairment induced by oxidative stress via promotion of antioxidant defense system. RSC Adv. 2017, 7, 36149–36162. [Google Scholar] [CrossRef]
- Qin, W.; Li, X.; Xie, L.; Li, S.; Liu, J.; Jia, L.; Dong, X.; Ren, X.; Xiao, J.; Yang, C. A long non-coding RNA, APOA4-AS, regulates APOA4 expression depending on HuR in mice. Nucleic Acids Res. 2016, 44, 6423–6433. [Google Scholar] [CrossRef]
- Qi, G.; Yang, L.; Xiao, C.; Shi, J.; Mi, Y.; Liu, X. Nutrient values and bioactivities of the extracts from three fern species in China: A comparative assessment. Food Funct. 2015, 6, 2918–2929. [Google Scholar] [CrossRef]
- Mi, Y.; Qi, G.; Fan, R.; Ji, X.; Liu, Z.; Liu, X. EGCG ameliorates diet-induced metabolic syndrome associating with the circadian clock. Biochim. Biophys. Acta. 2017, 1863, 1575–1589. [Google Scholar] [CrossRef]
Sample Availability: Samples of the resveratrol are not available from the authors. |






| Forward Primer | Reverse Primer | |
|---|---|---|
| Clock | AAAATACTCTCTACTCATCTGCTGG | ATGGCTCCTTTGGGTCTATTG |
| Bmal1 | CTGGCTAGAGTGTATACGTTTGG | GGTCACCTCAAAGCGATTTTC |
| Cry1 | TTACACTATGCTCATGGCGAC | GTGCTCTGTCTCTGGACTTTAG |
| Per1 | ATTCCGCCTAACCCCGTATGTGACC | GTGTGCCGCGTAGTGAAAATCCTCTTGT |
| Per2 | CCCTTCCGCATGACGCCCTACCTG | GACCGCCCTTTCATCCACATCCTG |
| Reverb-αα | CATGGTGCTACTGTGTAAGGTGTGT | CACAGGCGTGCACTCCATAG |
| β-actin | TCCACCTTCCAGCAGATGTG | GCATTTGCGGTGGACGAT |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Wei, L.; Zhao, C.; Li, J.; Liu, Z.; Zhang, M.; Wang, Y. Resveratrol Maintains Lipid Metabolism Homeostasis via One of the Mechanisms Associated with the Key Circadian Regulator Bmal1. Molecules 2019, 24, 2916. https://doi.org/10.3390/molecules24162916
Li J, Wei L, Zhao C, Li J, Liu Z, Zhang M, Wang Y. Resveratrol Maintains Lipid Metabolism Homeostasis via One of the Mechanisms Associated with the Key Circadian Regulator Bmal1. Molecules. 2019; 24(16):2916. https://doi.org/10.3390/molecules24162916
Chicago/Turabian StyleLi, Jing, Liping Wei, Caicai Zhao, Junyi Li, Zhigang Liu, Min Zhang, and Yutang Wang. 2019. "Resveratrol Maintains Lipid Metabolism Homeostasis via One of the Mechanisms Associated with the Key Circadian Regulator Bmal1" Molecules 24, no. 16: 2916. https://doi.org/10.3390/molecules24162916
APA StyleLi, J., Wei, L., Zhao, C., Li, J., Liu, Z., Zhang, M., & Wang, Y. (2019). Resveratrol Maintains Lipid Metabolism Homeostasis via One of the Mechanisms Associated with the Key Circadian Regulator Bmal1. Molecules, 24(16), 2916. https://doi.org/10.3390/molecules24162916