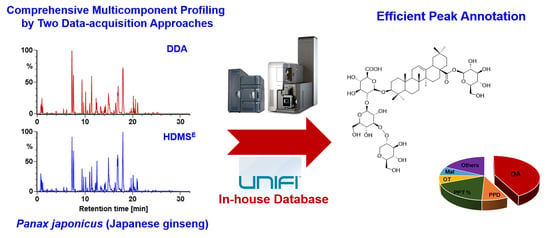
Integration of Data-Dependent Acquisition (DDA) and Data-Independent High-Definition MSE (HDMSE) for the Comprehensive Profiling and Characterization of Multicomponents from Panax japonicus by UHPLC/IM-QTOF-MS
Abstract
:1. Introduction
2. Results and Discussion
2.1. Optimization of the RP-UHPLC Conditions for Well Resolving the Multicomponents from P. japonicus
2.2. Optimization of the Parameters on Vion IM-QTOF Mass Spectrometer
2.3. Streamlined Workflows by Utilizing UNIFI to Automatedly Annotate both the DDA and HDMSE Data
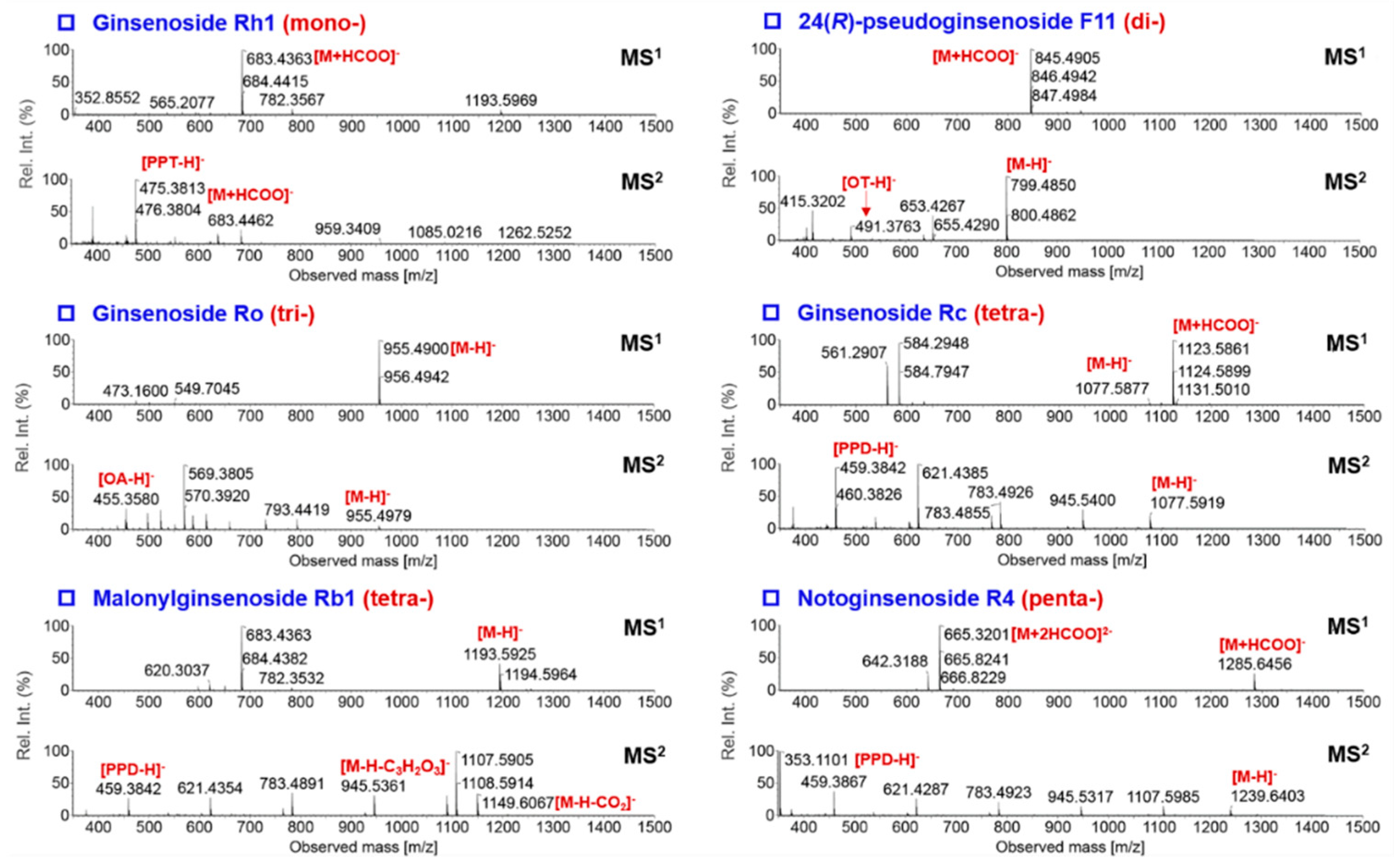
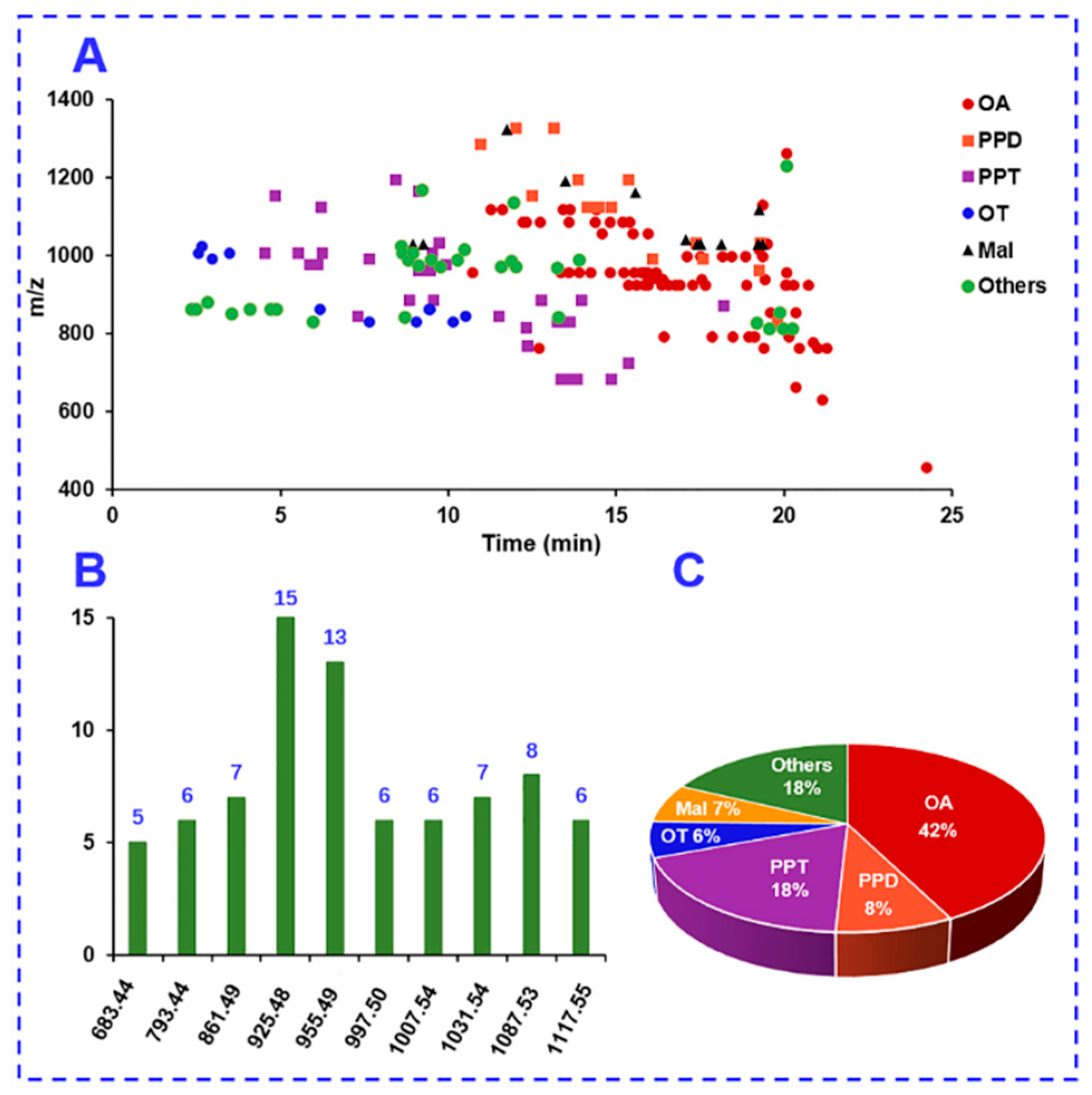
2.4. Complementarity of DDA and HDMSE for the Comprehensive Multicomponent Characterization
2.5. Comprehensive Characterization of Multicomponents from P. japonicus by Analyzing both the DDA and HDMSE Data
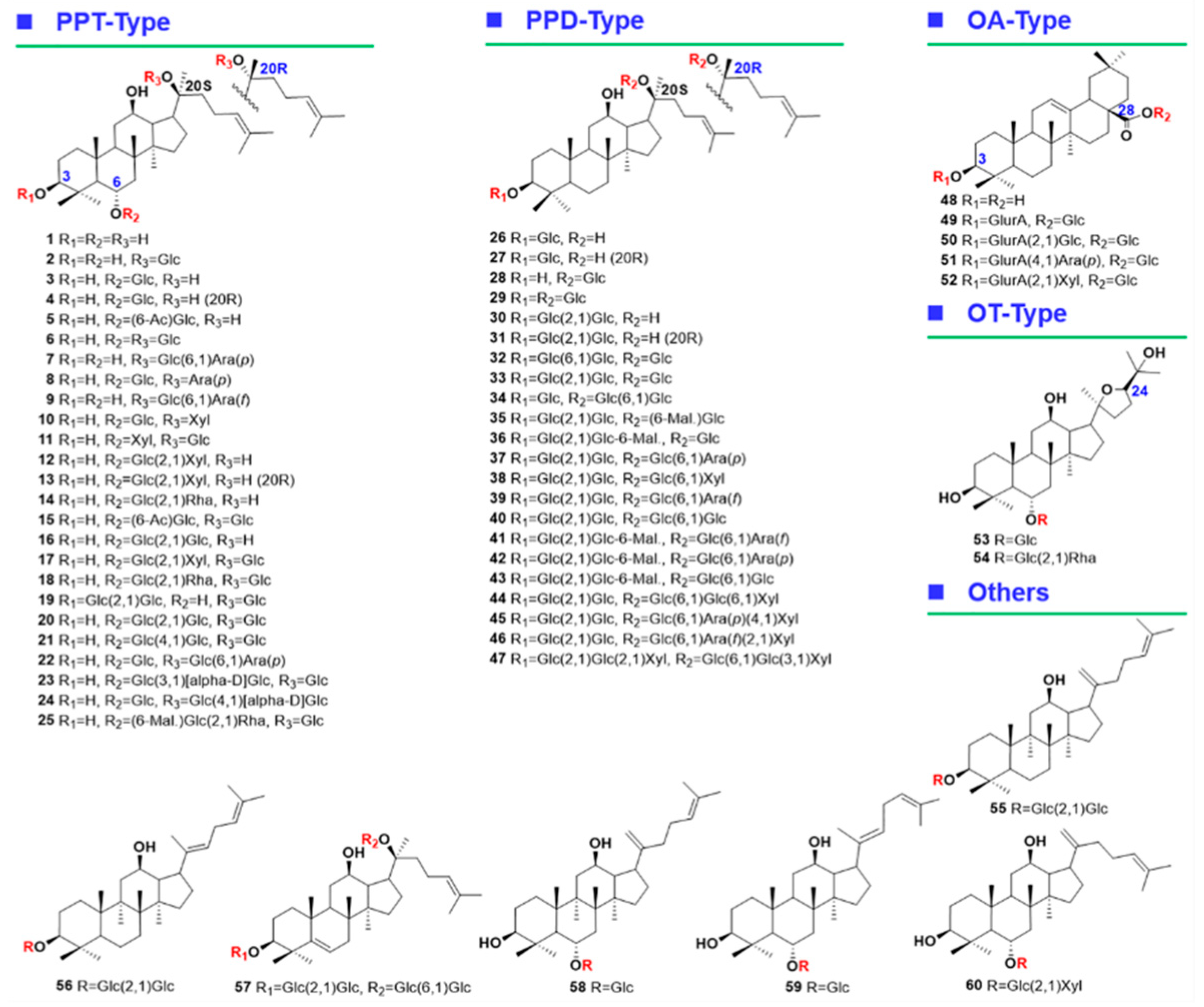
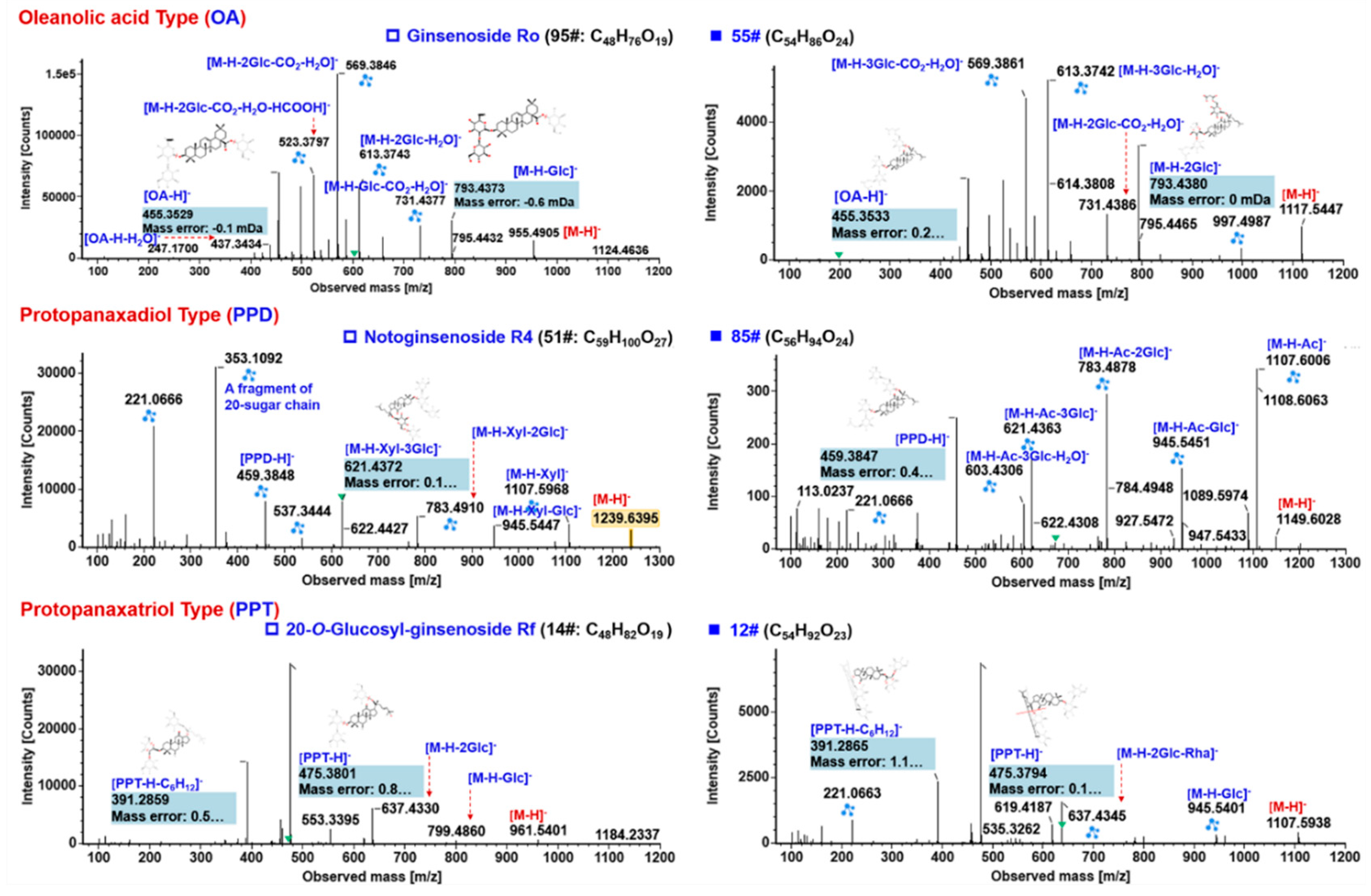
2.5.1. Characterization of OA-Ginsenosides Of the Panax Species
2.5.2. Characterization of PPD-/PPT-Ginsenosides
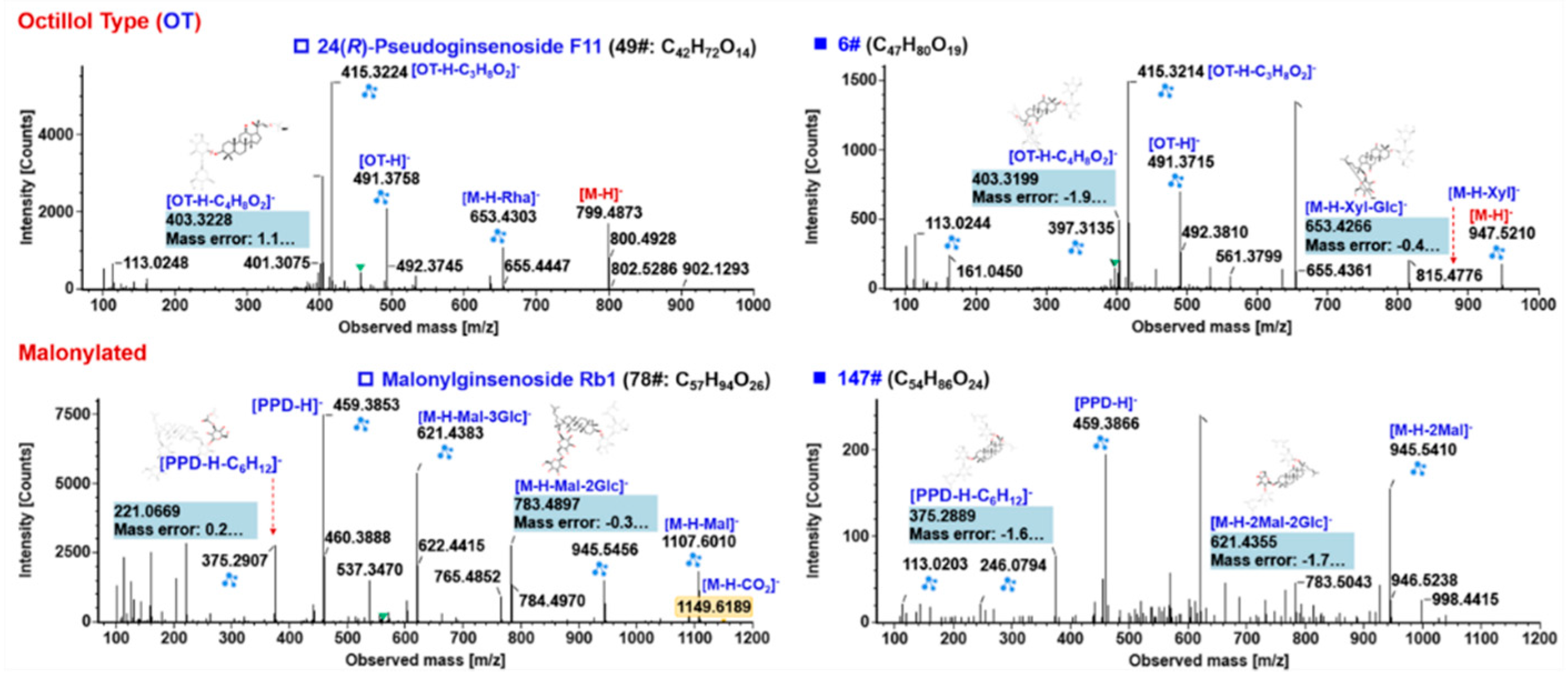
2.5.3. Characterization of OT-Ginsenosides
2.5.4. Characterization of Malonylated Ginsenosides
3. Materials and Methods
3.1. Reagents and Chemicals
3.2. Sample Preparation
3.3. UHPLC/IM-QTOF-MS
3.4. Date Processing
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Guo, D.A.; Wu, W.Y.; Ye, M.; Liu, X.; Cordell, G.A. A holistic approach to the quality control of traditional Chinese medicines. Science 2015, 347, S29–S31. [Google Scholar]
- Wolfender, J.L.; Marti, G.; Thomas, A.; Bertrand, S. Current approaches and challenges for the metabolite profiling of complex natural extracts. J. Chromatogr. A 2015, 1382, 136–164. [Google Scholar] [CrossRef] [PubMed]
- Jia, L.; Fu, L.L.; Wang, X.Y.; Yang, W.Z.; Wang, H.D.; Zuo, T.T.; Zhang, C.X.; Hu, Y.; Gao, X.M.; Han, L.F. Systematic profiling of the multicomponents and authentication of Erzhi Pill by UHPLC/Q-Orbitrap-MS oriented rapid polarity-switching data-dependent acquisition and selective monitoring of the chemical markers deduced from fingerprint analysis. Molecules 2018, 23, 3143. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.Z.; Ye, M.; Qiao, X.; Liu, C.F.; Miao, W.J.; Bo, T.; Tao, H.Y.; Guo, D.A. A strategy for efficient discovery of new natural compounds by integrating orthogonal column chromatography and liquid chromatography/mass spectrometry analysis: Its application in Panax ginseng, Panax quinquefolium and Panax notoginseng to characterize 437 potential new ginsenosides. Anal. Chim. Acta 2012, 739, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.X.; Zhang, C.X.; Hu, Y.; Zhang, M.H.; Wang, Y.N.; Qian, Y.X.; Yang, J.; Yang, W.Z.; Jiang, M.M.; Guo, D.A. Application of multiple chemical and biological approaches for quality assessment of Carthamus tinctorius L. (safflower) by determining both the primary and secondary metabolites. Phytomedicine 2019, 58, 152826. [Google Scholar] [CrossRef]
- Xie, W.W.; Jin, Y.R.; Hou, L.D.; Ma, Y.H.; Xu, H.J.; Zhang, K.R.; Zhang, L.T.; Du, Y.F. A practical strategy for the characterization of ponicidin metabolites in vivo and in vitro by UHPLC-Q-TOF-MS based on nontargeted SWATH data acquisition. J. Pharm. Biomed. Anal. 2017, 145, 865–878. [Google Scholar] [CrossRef]
- Xia, Y.G.; Gong, F.Q.; Guo, X.D.; Song, Y.; Li, C.X.; Liang, J.; Yang, B.Y.; Kuang, H.X. Rapid screening and characterization of triterpene saponins in Acanthopanax senticosus leaves via untargeted MSAll and SWATH techniques on a quadrupole time of flight mass spectrometry. J. Pharm. Biomed. Anal. 2019, 170, 68–82. [Google Scholar] [CrossRef]
- Plumb, R.S.; Johnson, K.A.; Rainville, P.; Smith, B.W.; Wilson, I.D.; Castro-Perez, J.M.; Nicholson, J.K. UPLC/MSE: A new approach for generating molecular fragment information for biomarker structure elucidation. Rapid Commun. Mass Spectrom. 2006, 20, 1989–1994. [Google Scholar] [CrossRef]
- Zhu, H.L.; Lin, H.Q.; Tan, J.; Wang, C.Z.; Wang, H.; Wu, F.L.; Dong, Q.H.; Liu, Y.H.; Li, P.Y.; Liu, J.P. UPLC-QTOF/MS-based nontargeted metabolomic analysis of mountain- and garden-cultivated Ginseng of different ages in Northeast China. Molecules 2019, 24, 33. [Google Scholar] [CrossRef]
- Naz, S.; Gallart-Ayala, H.; Reinke, S.N.; Mathon, C.; Blankley, R.; Chaleckis, R.; Wheelock, C.E. Development of a liquid chromatography-high resolution mass spectrometry metabolomics method with high specificity for metabolite identification using all ion fragmentation acquisition. Anal. Chem. 2017, 89, 7933–7942. [Google Scholar] [CrossRef]
- Navarro-Reig, M.; Jaumot, J.; Pina, B.; Moyano, E.; Galceran, M.T.; Tauler, R. Metabolomic analysis of the effects of cadmium and copper treatment in Oryza sativa L. using untargeted liquid chromatography coupled to high resolution mass spectrometry and all-ion fragmentation. Metabolomics 2017, 9, 660–675. [Google Scholar] [CrossRef] [PubMed]
- Kinyua, J.; Negreira, N.; Ibanez, M.; Bijlsma, L.; Hernandez, F.; Covaci, A.; van Nuijs, A.L.N. A data-independent acquisition workflow for qualitative screening of new psychoactive substances in biological samples. Anal. Bioanal. Chem. 2015, 407, 8773–8785. [Google Scholar] [CrossRef] [PubMed]
- Pettit, M.E.; Donnarumma, F.; Murray, K.K.; Solouki, T. Infrared laser ablation sampling coupled with data independent high resolution UPLC-MS/MS for tissue analysis. Anal. Chim. Acta 2018, 1034, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Qiu, S.; Yang, W.Z.; Yao, C.L.; Qiu, Z.D.; Shi, X.J.; Zhang, J.X.; Hou, J.J.; Wang, Q.R.; Wu, W.Y.; Guo, D.A. Nontargeted metabolomic analysis and “commercial-homophyletic” comparison-induced biomarkers verification for the systematic chemical differentiation of five different parts of Panax ginseng. J. Chromatogr. A 2016, 1453, 78–87. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Cai, Y.P.; Guo, Y.; Chen, F.F.; Zhu, Z.J. MetDIA: Targeted metabolite extraction of multiplexed MS/MS spectra generated by data-independent acquisition. Anal. Chem. 2016, 88, 8757–8764. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Jiang, J.M.; Zheng, D.; Yuan, M.; Wang, Z.Y.; Zhang, H.M.; Zhang, C.W.; Xiao, L.B.; Xu, H.X. A multidimensional analytical approach based on time-decoupled online comprehensive two-dimensional liquid chromatography coupled with ion mobility quadrupole time-of-flight mass spectrometry for the analysis of ginsenosides from white and red ginseng. J. Pharm. Biomed. Anal. 2019, 163, 24–33. [Google Scholar] [CrossRef]
- Jia, L.; Zuo, T.T.; Zhang, C.X.; Li, W.W.; Wang, H.D.; Hu, Y.; Wang, X.Y.; Qian, Y.X.; Yang, W.Z.; Yu, H.S. Simultaneous profiling and holistic comparison of the metabolomes among the flower buds of Panax ginseng, Panax quinquefolius, and Panax notoginseng by UHPLC/IM-QTOF-HDMSE-based metabolomics analysis. Molecules 2019, 34, 2188. [Google Scholar] [CrossRef]
- Shi, X.J.; Yang, W.Z.; Qiu, S.; Hou, J.J.; Wu, W.Y.; Guo, D.A. Systematic profiling and comparison of the lipidomes from Panax ginseng, P. quinquefolius, and P. notoginseng by ultrahigh performance supercritical fluid chromatography/high-resolution mass spectrometry and ion mobility-derived collision cross section measurement. J. Chromatogr. A 2018, 1548, 64–75. [Google Scholar] [CrossRef]
- Yang, W.Z.; Hu, Y.; Wu, W.Y.; Ye, M.; Guo, D.A. Saponins in the genus Panax L. (Araliaceae): A systematic review of their chemical diversity. Phytochemistry 2014, 106, 7–24. [Google Scholar] [CrossRef]
- Kang, S.; Min, H. Ginseng, the ‘immunity boost’: The effects of Panax ginseng on immune system. J. Ginseng Res. 2012, 36, 354–368. [Google Scholar] [CrossRef]
- Wang, T.; Guo, R.X.; Zhou, G.H.; Zhou, X.D.; Kou, Z.Z.; Sui, F.; Li, C.; Ling, L.Y.; Wang, Z.J. Traditional uses, botany, phytochemistry, pharmacology and toxicology of Panax notoginseng (Burk.) F.H. Chen: A review. J. Ethnopharmacol. 2016, 188, 234–258. [Google Scholar] [CrossRef] [PubMed]
- Qiu, S.; Yang, W.Z.; Shi, X.J.; Yao, C.L.; Yang, M.; Liu, X.; Jiang, B.H.; Wu, W.Y.; Guo, D.A. A green protocol for efficient discovery of novel natural compounds: Characterization of new ginsenosides from the stem and leaves of Panax ginseng. Anal. Chim. Acta 2015, 893, 65–76. [Google Scholar] [CrossRef] [PubMed]
- National Pharmacopeia Commission. Pharmacopoeia of the People’s Republic of China, Part I; China Medical Science Press: Beijing, China, 2015; pp. 138–139. [Google Scholar]
- Meng, F.C.; Wu, Q.S.; Wang, R.B.; Li, S.P.; Lin, L.G.; Chen, P.; Zhang, Q.W. A novel strategy for quantitative analysis of major ginsenosides in Panacis Japonici Rhizoma with a standardized reference fraction. Molecules 2017, 22, 2067. [Google Scholar] [CrossRef] [PubMed]
- He, Y.M.; Ai, K.; He, C.X.; Zhang, C.C.; Yuan, D.; Cai, S.J. Triterpene saponins in Panax japonicus and their 13C-NMR spectroscopic characteristics. Chin. J. Chin. Mater. Med. 2019, 44, 249–260. [Google Scholar]
- Chen, J.L.; Tan, M.X.; Zou, L.S.; Liu, X.H.; Chen, S.Y.; Shi, J.J.; Wang, C.C.; Mei, Y.Q. Simultaneous determination of multiple bioactive constituents in Panacis Japonici Rhizoma processed by different methods and grey relational analysis. Chin. J. Chin. Mater. Med. 2018, 43, 4274–4282. [Google Scholar]
- Chen, J.L.; Tan, M.X.; Zou, L.S.; Liu, X.H.; Chen, S.Y.; Shi, J.J.; Wang, C.C.; Mei, Y.Q. Study on Saponins in Panax Japonici Rhizoma by UFLC-triple TOF MS/MS. Food Sci. 2019, 1–16. [Google Scholar]
- Du, Z.X.; Li, J.H.; Zhang, X.; Pei, J.; Huang, L.F. An integrated LC-MS-based strategy for the quality assessment and discrimination of three Panax species. Molecules 2018, 23, 2988. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.Z.; Qiao, X.; Li, K.; Fan, J.R.; Bo, T.; Guo, D.A.; Ye, M. Identification and differentiation of Panax ginseng, Panax quinquefolium, and Panax notoginseng by monitoring multiple diagnostic chemical markers. Acta Pharm. Sin. B 2016, 6, 568–575. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.J.; Yang, W.Z.; Qiu, S.; Yao, C.L.; Shen, Y.; Pan, H.Q.; Bi, Q.R.; Yang, M.; Wu, W.Y.; Guo, D.A. An in-source multiple collision-neutral loss filtering based nontargeted metabolomics approach for the comprehensive analysis of malonyl-ginsenosides from Panax ginseng, P. quinquefolius, and P. notoginseng. Anal. Chim. Acta 2017, 952, 59–70. [Google Scholar] [CrossRef]
- Fu, L.L.; Ding, H.; Han, L.F.; Jia, L.; Yang, W.Z.; Zhang, C.X.; Hu, Y.; Zuo, T.T.; Gao, X.M.; Guo, D.A. Simultaneously targeted and untargeted multicomponent characterization of Erzhi Pill by offline two-dimensional liquid chromatography/quadrupole-Orbitrap mass spectrometry. J. Chromatogr. A 2019, 1584, 87–96. [Google Scholar] [CrossRef]
- Si, W.; Yang, W.Z.; Guo, D.A.; Wu, J.; Zhang, J.X.; Qiu, S.; Yao, C.L.; Cui, Y.J.; Wu, W.Y. Selective ion monitoring of quinochalcone C-glycoside markers for the simultaneous identification of Carthamus tinctorius L. in eleven Chinese patent medicines by UHPLC/QTOF MS. J. Pharm. Biomed. Anal. 2016, 117, 510–521. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.Z.; Bo, T.; Ji, S.; Qiao, X.; Guo, D.A.; Ye, M. Rapid chemical profiling of saponins in the flower buds of Panax notoginseng by integrating MCI gel column chromatography and liquid chromatography/mass spectrometry analysis. Food Chem. 2013, 139, 762–769. [Google Scholar] [CrossRef]
- Samimi, R.; Xu, W.Z.; Lui, E.M.K.; Charpentier, P.A. Isolation and immunosuppressive effects of 6”-O-acetylginsenoside Rb1 extracted from North American Ginseng. Planta Med. 2014, 80, 509–516. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Xu, Y.R.; Yang, J.J.; Wang, W.Z.; Zhang, J.Q.; Zhang, R.M.; Meng, Q.G. Discovery, semisynthesis, biological activities, and metabolism of octillol-type saponins. J. Ginseng Res. 2017, 41, 373–378. [Google Scholar] [CrossRef] [PubMed]
- Duc, N.M.; Dasai, R.; Ohtani, K.; Ito, A.; Nham, N.T.; Yamasaki, K.; Tanaka, O. Saponins from Vietnamese ginseng, Panax vietnamensis Ha et Grushv. Collected in central Vietnam. II. Chem. Pharm. Bull. 1994, 42, 115–122. [Google Scholar]
- Qiu, S.; Yang, W.Z.; Yao, C.L.; Shi, X.J.; Li, J.Y.; Lou, Y.; Duan, Y.N.; Wu, W.Y.; Guo, D.A. Malonylginsenosides with potential antidiabetic activities from the flower buds of Panax ginseng. J. Nat. Prod. 2017, 80, 899–908. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Li, W.; Li, X.; Zhang, M.; Chen, L.; Zhang, Y.N.; Sun, G.Z.; Ruan, C.C. Antidiabetic effects of malonylginsenosides from Panax ginseng on type 2 diabetic rats induced by high-fat diet and streptozotocin. J. Ethnopharmacol. 2013, 145, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.J.; Yang, W.Z.; Huang, Y.; Hou, J.J.; Qiu, S.; Yao, C.L.; Feng, Z.J.; Wei, W.L.; Wu, W.Y.; Guo, D.A. Direct screening of malonylginsenosides from nine Ginseng extracts by an untargeted profiling strategy incorporating in-source collision-induced dissociation, mass tag, and neutral loss scan on a hybrid linear ion-trap/Orbitrap mass spectrometer coupled to ultra-high performance liquid chromatography. J. Chromatogr. A 2018, 1571, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Paglia, G.; Angel, P.; Williams, J.P.; Richardson, K.; Olivos, H.J.; Thompson, J.W.; Menikarachchi, L.; Lai, S.; Walsh, C.; Moseley, A.; et al. Ion mobility-derived collision cross section as an additional measure for lipid fingerprinting and identification. Anal. Chem. 2015, 87, 1137–1144. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Sample of Panax japonicus is available from the authors. |



© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, C.; Zuo, T.; Wang, X.; Wang, H.; Hu, Y.; Li, Z.; Li, W.; Jia, L.; Qian, Y.; Yang, W.; et al. Integration of Data-Dependent Acquisition (DDA) and Data-Independent High-Definition MSE (HDMSE) for the Comprehensive Profiling and Characterization of Multicomponents from Panax japonicus by UHPLC/IM-QTOF-MS. Molecules 2019, 24, 2708. https://doi.org/10.3390/molecules24152708
Zhang C, Zuo T, Wang X, Wang H, Hu Y, Li Z, Li W, Jia L, Qian Y, Yang W, et al. Integration of Data-Dependent Acquisition (DDA) and Data-Independent High-Definition MSE (HDMSE) for the Comprehensive Profiling and Characterization of Multicomponents from Panax japonicus by UHPLC/IM-QTOF-MS. Molecules. 2019; 24(15):2708. https://doi.org/10.3390/molecules24152708
Chicago/Turabian StyleZhang, Chunxia, Tiantian Zuo, Xiaoyan Wang, Hongda Wang, Ying Hu, Zheng Li, Weiwei Li, Li Jia, Yuexin Qian, Wenzhi Yang, and et al. 2019. "Integration of Data-Dependent Acquisition (DDA) and Data-Independent High-Definition MSE (HDMSE) for the Comprehensive Profiling and Characterization of Multicomponents from Panax japonicus by UHPLC/IM-QTOF-MS" Molecules 24, no. 15: 2708. https://doi.org/10.3390/molecules24152708