Dexamethasone-Induced Mitochondrial Dysfunction and Insulin Resistance-Study in 3T3-L1 Adipocytes and Mitochondria Isolated from Mouse Liver
Abstract
:1. Introduction
2. Results
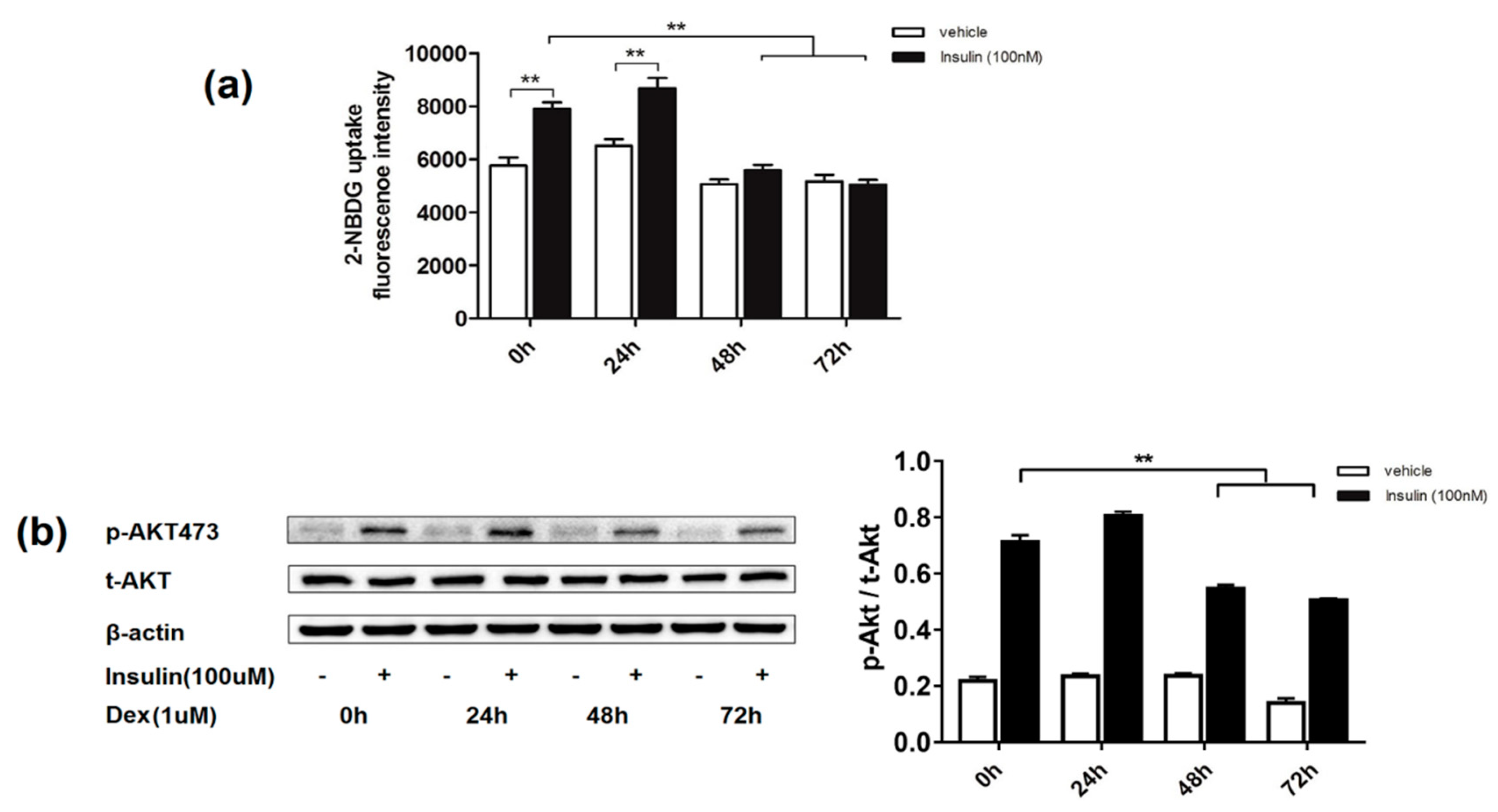
2.1. Effects of Dexamethasone on Insulin-Induced Glucose Uptake and AKT Phosphorylation Level in 3T3-L1 Adipocytes
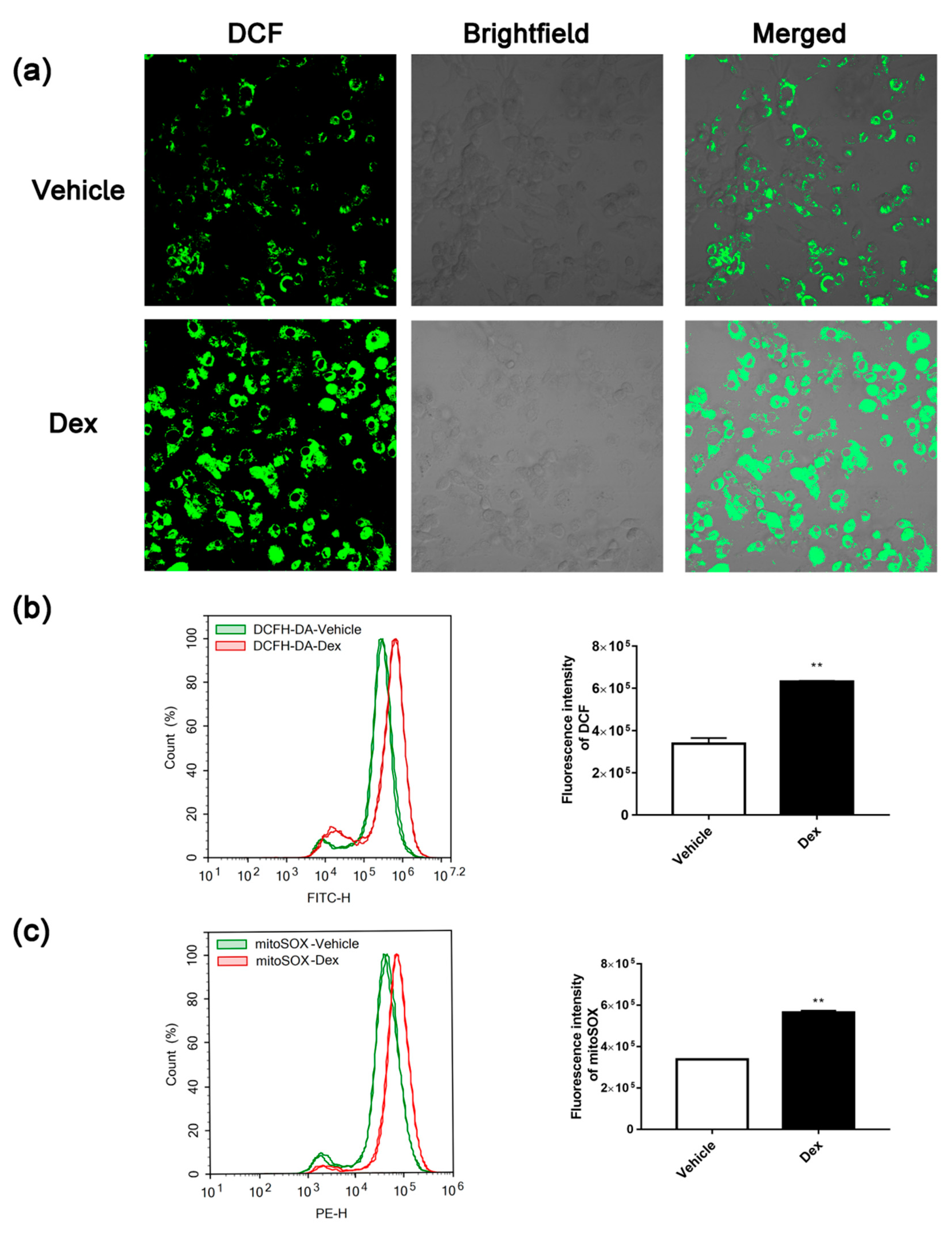
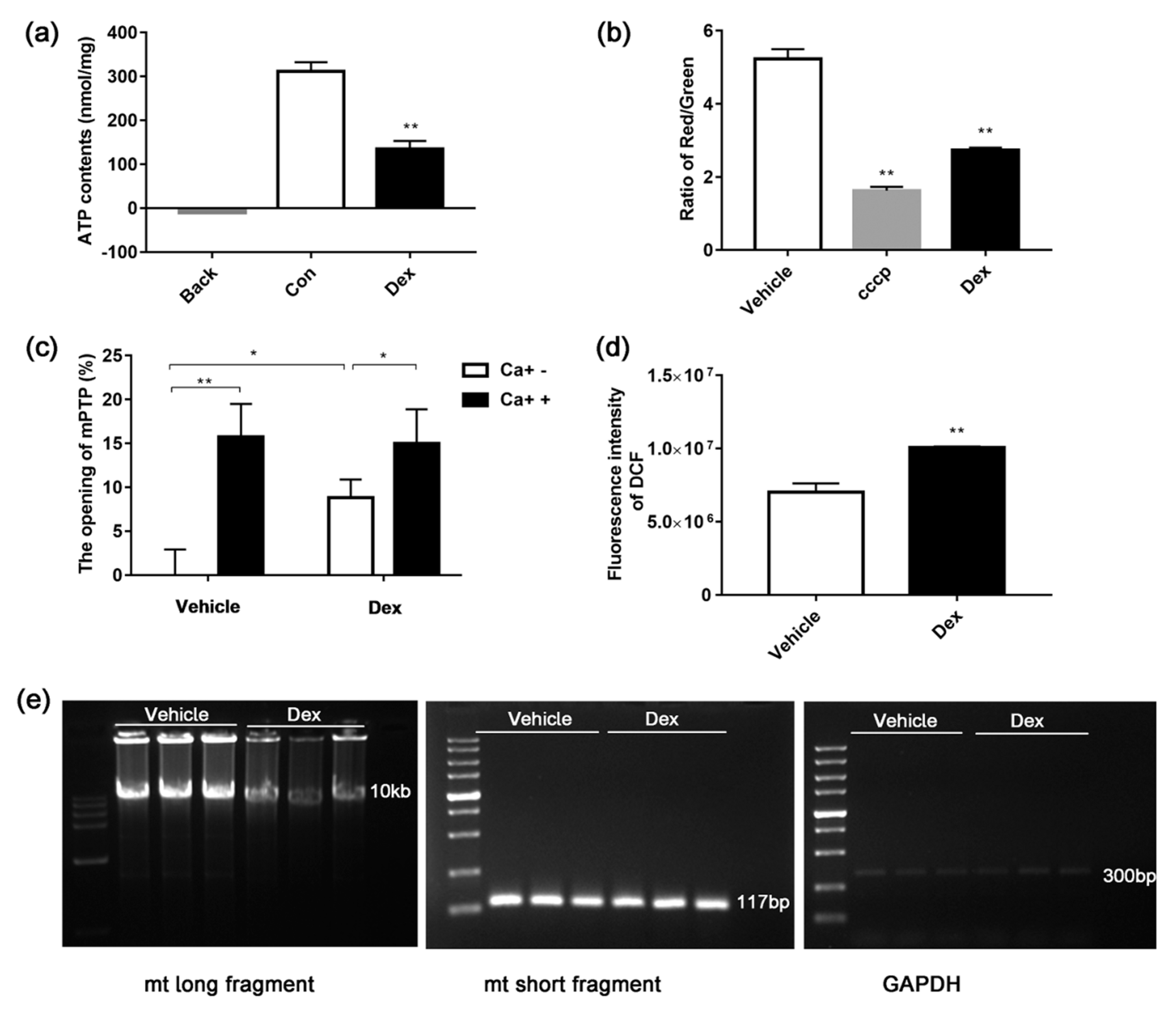
2.2. Dexamethasone Treatment-Induced ROS Accumulation
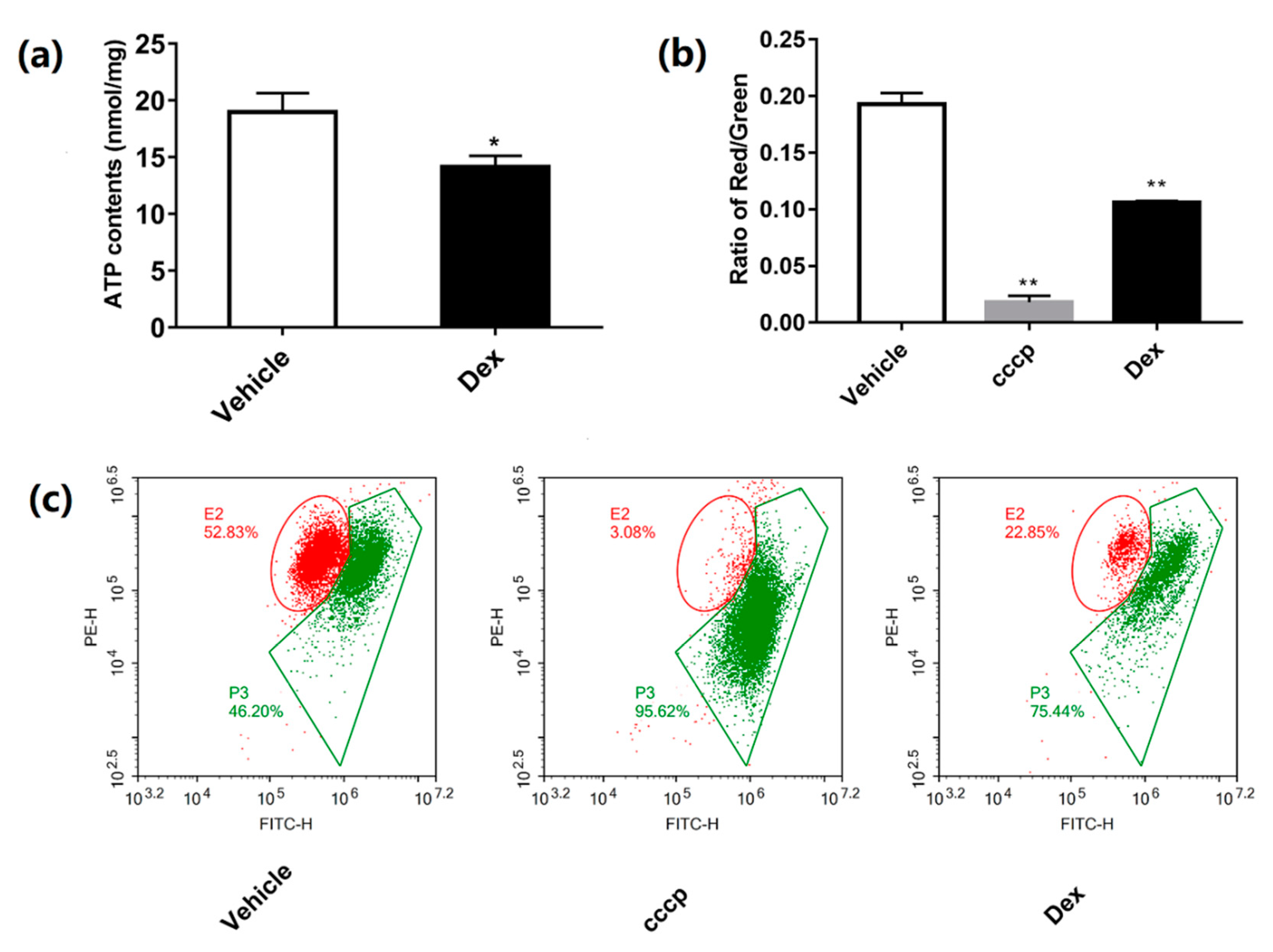
2.3. Dexamethasone Reduces MMP and ATP Production
2.4. Effects of Dexamethasone on Mitochondrial Mass and mtDNA
2.5. Dexamethasone Treatment Affects Mitochondrial Biogenesis and Dynamics in 3T3-L1 Adipocytes
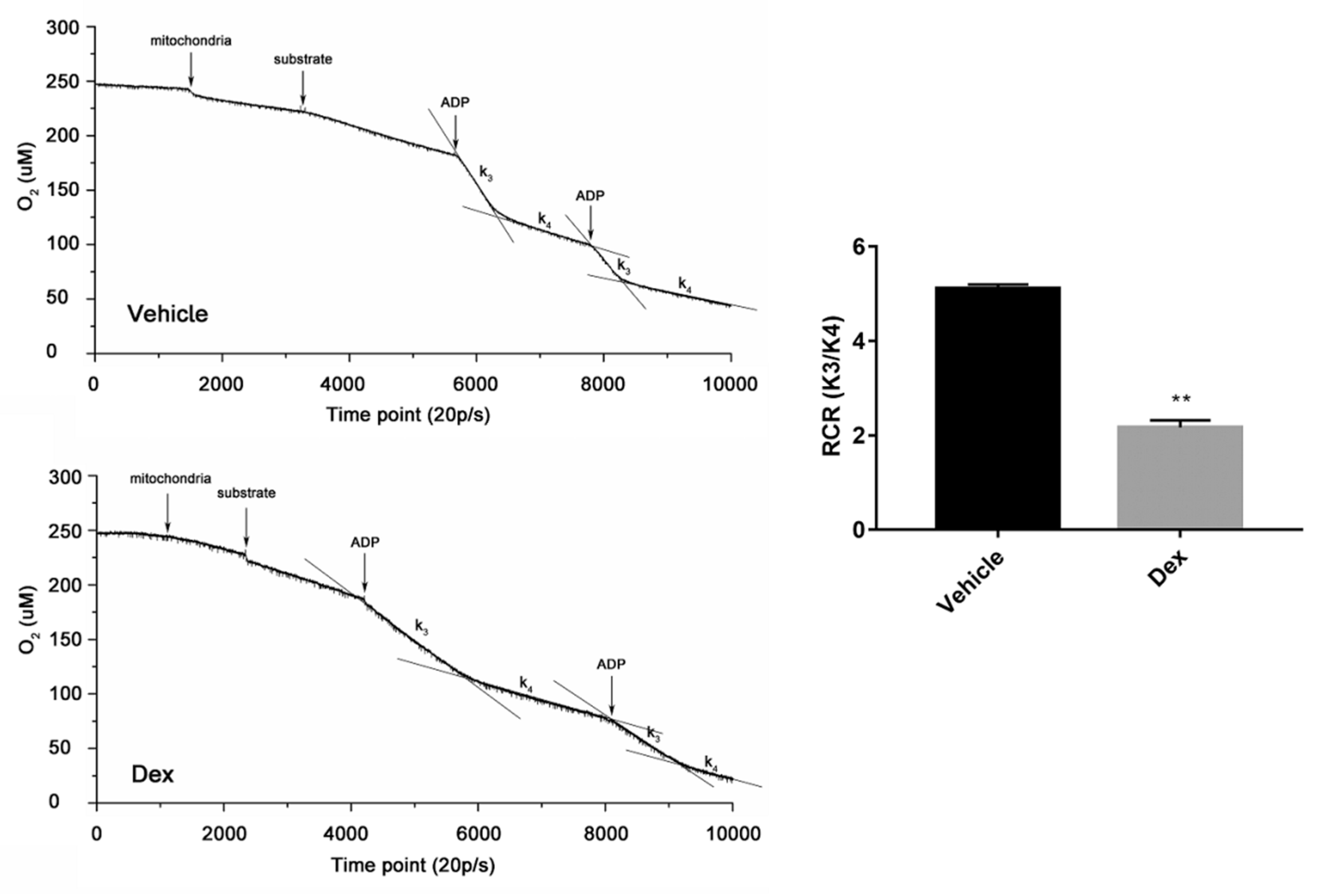
2.6. Effects of Dexamethasone on Mitochondria Isolated from Mouse Liver
3. Discussion
4. Materials and Methods
4.1. 3T3-L1 Cell Culture and Induction of Insulin Resistance
4.2. Experimental Animals
4.3. Preparation of Liver Mitochondria Suspension
4.4. 2-NBDG Uptake Assay
4.5. Western Blot Analysis
4.6. Determination of ROS Generation
4.7. ATP Content Measurement
4.8. MMP Detection
4.9. Mitochondria Mass
4.10. DNA and RNA Extraction and cDNA Synthesis
4.11. Real-time Quantitative PCR (RT-PCR)
4.12. Long PCR Experiments
4.13. Isolated Mitochondrial Respiratory Test
4.14. Determination of Opening of mPTP
4.15. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Steinberger, J.; Daniels, S.R. Obesity, Insulin Resistance, Diabetes, and Cardiovascular Risk in Children. Circulation 2003, 107, 1448–1453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saad, M.F.; Rewers, M.; Selby, J.; Howard, G.; Jinagouda, S.; Fahmi, S.; Zaccaro, D.; Bergman, R.N.; Savage, P.J.; Haffner, S.M. Insulin resistance and hypertension: The Insulin Resistance Atherosclerosis study. Hypertension 2004, 43, 1324–1331. [Google Scholar] [CrossRef] [PubMed]
- Owen, O.E.; Cahill, G.F. Metabolic Effects of Exogenous Glucocorticoids in Fasted Man. J. Clin. Investig. 1973, 52, 2596–2605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sangeetha, K.N.; Shilpa, K.; Kumari, P.J.; Lakshmi, B.S. Reversal of dexamethasone induced insulin resistance in 3T3L1 adipocytes by 3beta-taraxerol of Mangifera indica. Phytomedicine 2013, 20, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Olefsky, J.M.; Johnson, J.; Liu, F.; Jen, P.; Reaven, G.M. The effects of acute and chronic dexamethasone administration on insulin binding to isolated rat hepatocytes and adipocytes. Metab. Clin. Exp. 1975, 24, 517–527. [Google Scholar] [CrossRef]
- Haber, R.S.; Weinstein, S.P. Role of glucose transporters in glucocorticoid-induced insulin resistance. GLUT4 isoform in rat skeletal muscle is not decreased by dexamethasone. Diabetes 1992, 41, 728–735. [Google Scholar] [CrossRef]
- Shyni, G.L.; Sasidharan, K.; Francis, S.K.; Das, A.A.; Nair, M.S.; Raghu, K.G. Licarin B from Myristica fragrans improves insulin sensitivity via PPARγ and activation of GLUT4 in the IRS-1/PI3K/AKT pathway in 3T3-L1 adipocytes. RSC Adv. 2016, 6, 79859–79870. [Google Scholar] [CrossRef]
- Pan, X.; Zaarur, N.; Singh, M.; Morin, P.; Kandror, K.V.; Brennwald, P.J. Sortilin and retromer mediate retrograde transport of Glut4 in 3T3-L1 adipocytes. Mol. Boil. Cell 2017, 28, 1667–1675. [Google Scholar] [CrossRef]
- Sakoda, H.; Ogihara, T.; Anai, M.; Funaki, M.; Inukai, K.; Katagiri, H.; Fukushima, Y.; Onishi, Y.; Ono, H.; Fujishiro, M.; et al. Dexamethasone-induced insulin resistance in 3T3-L1 adipocytes is due to inhibition of glucose transport rather than insulin signal transduction. Diabetes 2000, 49, 1700–1708. [Google Scholar] [CrossRef]
- Yuzefovych, L.V.; Musiyenko, S.I.; Wilson, G.L.; Rachek, L.I. Mitochondrial DNA Damage and Dysfunction, and Oxidative Stress Are Associated with Endoplasmic Reticulum Stress, Protein Degradation and Apoptosis in High Fat Diet-Induced Insulin Resistance Mice. PLOS ONE 2013, 8, e54059. [Google Scholar] [CrossRef]
- Ramalingam, L.; Menikdiwela, K.; Lemieux, M.; Dufour, J.M.; Kaur, G.; Kalupahana, N.; Moustaid-Moussa, N.; Menikdewella, K. The renin angiotensin system, oxidative stress and mitochondrial function in obesity and insulin resistance. Biochim. et Biophys. Acta (BBA) Mol. Basis 2017, 1863, 1106–1114. [Google Scholar] [CrossRef]
- Wilson-Fritch, L.; Nicoloro, S.; Chouinard, M.; Lazar, M.A.; Chui, P.C.; Leszyk, J.; Straubhaar, J.; Czech, M.P.; Corvera, S. Mitochondrial remodeling in adipose tissue associated with obesity and treatment with rosiglitazone. J. Clin. Investig. 2004, 114, 1281–1289. [Google Scholar] [CrossRef] [PubMed]
- Kelley, D.E.; He, J.; Menshikova, E.V.; Ritov, V.B. Dysfunction of Mitochondria in Human Skeletal Muscle in Type 2 Diabetes. Diabetes 2002, 51, 2944–2950. [Google Scholar] [CrossRef] [Green Version]
- Petersen, K.F.; Dufour, S.; Befroy, D.; Garcia, R.; Shulman, G.I. Impaired Mitochondrial Activity in the Insulin-Resistant Offspring of Patients with Type 2 Diabetes. New Engl. J. Med. 2004, 350, 664–671. [Google Scholar] [CrossRef]
- St-Pierre, J.; Brand, M.D.; Boutilier, R.G. Mitochondria as ATP consumers: Cellular treason in anoxia. Proc. Acad. Sci. 2000, 97, 8670–8674. [Google Scholar] [CrossRef] [Green Version]
- Robinson, E.D.; Wong, R. Mitochondria1 DNA Molecules and Virtual Number of Mitochondria per Cell in Mammalian Cells. J Cell Physiol. 1988, 3, 507–513. [Google Scholar] [CrossRef] [PubMed]
- Choksi, K.; Boylston, W.; Rabek, J.; Widger, W.; Papaconstantinou, J. Oxidatively damaged proteins of heart mitochondrial electron transport complexes. Biochim. et Biophys. Acta (BBA) Mol. Basis 2004, 1688, 95–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, F.B.; Manson, J.A.E.; Stampfer, M.J.; Colditz, G.; Liu, S.; Solomon, C.G.; Willett, W.C. Diet, lifestyle, and the risk of type 2 diabetes mellitus in women. N. Engl. J. Med. 2001, 345, 790–797. [Google Scholar] [CrossRef] [PubMed]
- Hajer, G.R.; Van Haeften, T.W.; Visseren, F.L. Adipose tissue dysfunction in obesity, diabetes, and vascular diseases. Eur. Hear. J. 2008, 29, 2959–2971. [Google Scholar] [CrossRef] [Green Version]
- Qin, J.-H.; Ma, J.-Z.; Yang, X.-W.; Hu, Y.-J.; Zhou, J.; Fu, L.-C.; Tian, R.-H.; Liu, S.; Xu, G.; Shen, X.-L. A Triterpenoid Inhibited Hormone-Induced Adipocyte Differentiation and Alleviated Dexamethasone-Induced Insulin Resistance in 3T3-L1 adipocytes. Nat. Prod. Bioprospect. 2015, 5, 159–166. [Google Scholar] [CrossRef] [Green Version]
- Kaaman, M.; Sparks, L.M.; Van Harmelen, V.; Smith, S.R.; Sjolin, E.; Dahlman, I.; Arner, P.; Harmelen, V. Strong association between mitochondrial DNA copy number and lipogenesis in human white adipose tissue. Diabetologia 2007, 50, 2526–2533. [Google Scholar] [CrossRef] [PubMed]
- Limón-Pacheco, J.; Gonsebatt, M.E. The role of antioxidants and antioxidant-related enzymes in protective responses to environmentally induced oxidative stress. Res. Toxicol. Environ. Mutagen. 2009, 674, 137–147. [Google Scholar] [CrossRef]
- Montier, L.L.C.; Deng, J.J.; Bai, Y. Number matters: control of mammalian mitochondrial DNA copy number. J. Genet. Genom. 2009, 36, 125–131. [Google Scholar] [CrossRef] [Green Version]
- Zong, H.; Ren, J.M.; Young, L.H.; Pypaert, M.; Mu, J.; Birnbaum, M.J.; Shulman, G.I. AMP kinase is required for mitochondrial biogenesis in skeletal muscle in response to chronic energy deprivation. Proc. Acad. Sci. 2002, 99, 15983–15987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelly, D.P. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 2004, 18, 357–368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suen, D.-F.; Norris, K.L.; Youle, R.J. Mitochondrial dynamics and apoptosis. Genome Res. 2008, 22, 1577–1590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Brito, O.M.; Scorrano, L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nat. Cell Boil. 2008, 456, 605–610. [Google Scholar] [CrossRef]
- Luan, G.; Wang, Y.; Wang, Z.; Zhou, W.; Hu, N.; Li, G.; Wang, H. Flavonoid Glycosides from Fenugreek Seeds Regulate Glycolipid Metabolism by Improving Mitochondrial Function in 3T3-L1 Adipocytes in Vitro. J. Agric. Food. Chem. 2018, 66, 3169–3178. [Google Scholar] [CrossRef]
- Houstis, N.; Rosen, E.D.; Lander, E.S. Reactive oxygen species have a causal role in multiple forms of insulin resistance. Nat. Cell Boil. 2006, 440, 944–948. [Google Scholar] [CrossRef]
- Yang, X.-X.; Xu, F.; Wang, D.; Yang, Z.-W.; Tan, H.-R.; Shang, M.-Y.; Wang, X.; Cai, S.-Q. Development of a mitochondria-based centrifugal ultrafiltration/liquid chromatography/mass spectrometry method for screening mitochondria-targeted bioactive constituents from complex matrixes: Herbal medicines as a case study. J. Chromatogr. A 2015, 1413, 33–46. [Google Scholar] [CrossRef]
- Luan, G.; Tie, F.; Yuan, Z.; Li, G.; He, J.; Wang, Z.; Wang, H. Hypaphorine, an Indole Alkaloid Isolated from Caragana korshinskii Kom., Inhibites 3T3-L1 Adipocyte Differentiation and Improves Insulin Sensitivity in Vitro. Chem. Biodivers. 2017, 14, e1700038. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.C.; Sun, G.B.; Wang, J.; Zhang, H.J.; Sun, X.B. Naringin protects against anoxia/reoxygenation-induced apoptosis in H9c2 cells via the Nrf2 signaling pathway. Food Funct. 2015, 6, 1331–1344. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Chen, D.; Xie, Q.; Yang, Y.; Shen, W. Nebivolol stimulates mitochondrial biogenesis in 3T3-L1 adipocytes. Biochem. Biophys. Commun. 2013, 438, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Nolan, T.; Hands, R.E.; Bustin, S.A. Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 2006, 1, 1559–1582. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Santos, J.H.; Mandavilli, B.S.; Houten, B.V. Measuring Oxidative mtDNA Damage and Repair Using Quantitative PCR. Methods Mol Biol. 2002, 197, 159–176. [Google Scholar]
- Sun, C.; Liu, X.; Di, C.; Wang, Z.; Mi, X.; Liu, Y.; Zhao, Q.; Mao, A.; Chen, W.; Gan, L.; et al. MitoQ Regulates Autophagy by Inducing a Pseudo-Mitochondrial Membrane Potential (PMMP). Autophagy 2017, 13, 730–738. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |


| Target Gene | Primer Sequence | Size (bp) | Accession Numbers |
|---|---|---|---|
| Mfn1 | Forward: 5′-GCTGTCAGAGCCCATCTTTC-3′ Reverse: 5′-CAGCCCACTGTTTTCCAAAT-3′ | 195 | NM_024200 |
| Mfn2 | Forward: 5′-GCCAGCTTCCTTGAAGACAC-3′ Reverse: 5′-GCAGAACTTTGTCCCAGAGC-3′ | 208 | NM_001355590 |
| DRP1 | Forward: 5′-ATGCCTGTGGGCTAATGAAC-3′ Reverse: 5′-AGTTGCCTGTTGTTGGTTCC-3′ | 180 | NM_001360010 |
| PGC-1α | Forward: 5′- CGGAAATCATATCCAACCAG-3′ Reverse: 5′-TGAGGACCGCTAGCAAGTTTG-3′ | 243 | XM_006503779 |
| NRF1 | Forward: 5′- TGGTCCAGAGAGTGCTTGTG-3′ Reverse: 5′- TTCCTGGGAAGGGAGAAGAT-3′ | 184 | NM_001361693 |
| TFam | Forward: 5′-GGAATGTGGAGCGTGCTAAAA-3′ Reverse: 5′-TGCTGGAAAAACACTTCGGAATA-3′ | 118 | NM_009360 |
| Long fragment | Forward: 5′-TACTAGTCCGCGAGCCTTCAAAGC-3′ Reverse: 5′-GGGTGATCTTTGTTTGCGGGT-3′ | 8636 | AJ512208.1 |
| Short fragment | Forward: 5′- CCCAGCTACTACCATCATTCAAGT -3′ Reverse: 5′-GATGGTTTGGGAGATTGGTTGATG -3′ | 117 | NC_005089 |
| β-actin | Forward: 5′-CCTGAGGCTCTTTTCCAGCC-3′ Reverse: 5′-TAGAGGTCTTTACGGATGTCAACGT-3′ | 110 | NM_007393 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luan, G.; Li, G.; Ma, X.; Jin, Y.; Hu, N.; Li, J.; Wang, Z.; Wang, H. Dexamethasone-Induced Mitochondrial Dysfunction and Insulin Resistance-Study in 3T3-L1 Adipocytes and Mitochondria Isolated from Mouse Liver. Molecules 2019, 24, 1982. https://doi.org/10.3390/molecules24101982
Luan G, Li G, Ma X, Jin Y, Hu N, Li J, Wang Z, Wang H. Dexamethasone-Induced Mitochondrial Dysfunction and Insulin Resistance-Study in 3T3-L1 Adipocytes and Mitochondria Isolated from Mouse Liver. Molecules. 2019; 24(10):1982. https://doi.org/10.3390/molecules24101982
Chicago/Turabian StyleLuan, Guangxiang, Gang Li, Xiao Ma, Youcai Jin, Na Hu, Ji Li, Zhenhua Wang, and Honglun Wang. 2019. "Dexamethasone-Induced Mitochondrial Dysfunction and Insulin Resistance-Study in 3T3-L1 Adipocytes and Mitochondria Isolated from Mouse Liver" Molecules 24, no. 10: 1982. https://doi.org/10.3390/molecules24101982






