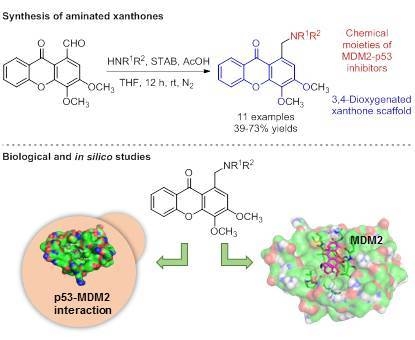
Synthesis, Biological Evaluation, and In Silico Studies of Novel Aminated Xanthones as Potential p53-Activating Agents
Abstract
:1. Introduction
2. Results and Discussion
2.1. Chemistry
2.1.1. Synthesis of 3,4-dimethoxy-9-oxo-9H-xanthene-1-carbaldehyde (5)
2.1.2. Synthesis of a Library of Aminated Xanthones (30–40)
2.2. Biological Activity Evaluation
2.2.1. Effect of Synthesized Xanthone Derivatives 30–40 on the MDM2-p53 Interaction Using A Yeast-Based Screening Assay
2.2.2. Evaluation of the Antitumor Activity of Xanthone 37 in Human Cancer Cell Lines
2.3. In Slico Studies
3. Materials and Methods
3.1. Chemistry
3.1.1. Synthesis of 3,4-dimethoxy-1-methyl-9H-xanthen-9-one (27)
3.1.2. Synthesis of 1-(dibromomethyl)-3,4-dimethoxy-9H-xanthen-9-one (28)
3.1.3. Synthesis of 3,4-dimethoxy-9-oxo-9H-xanthene-1-carbaldehyde (5)
3.1.4. Synthesis of 1-(hydroxymethyl)-3,4-dimethoxy-9H-xanthen-9-one (29)
3.1.5. General Procedure for the Synthesis of Xanthones (30–40)
3.2. Biological Evaluation
3.2.1. Yeast Strains and Growth Conditions
3.2.2. Human Cancer Cell Lines and Culture Conditions
3.2.3. Cell Proliferation Assay
3.2.4. Colony Formation Assay
3.2.5. Cell Cycle Analysis
3.2.6. Western Blot Analysis
3.2.7. Statistical Analysis
3.3. In Silico Studies onto MDM2
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Lane, D.P. Cancer. p53, guardian of the genome. Nature 1992, 358, 15–16. [Google Scholar] [CrossRef]
- Shaw, P.H. The role of p53 in cell cycle regulation. Pathol. Res. Pract. 1996, 192, 669–675. [Google Scholar] [CrossRef]
- Toledo, F.; Wahl, G.M. Regulating the p53 pathway: In vitro hypotheses, in vivo veritas. Nat. Rev. Cancer 2006, 6, 909–923. [Google Scholar] [CrossRef] [PubMed]
- Vazquez, A.; Bond, E.E.; Levine, A.J.; Bond, G.L. The genetics of the p53 pathway, apoptosis and cancer therapy. Nat. Rev. Drug Discov. 2008, 7, 979–987. [Google Scholar] [CrossRef] [PubMed]
- Rufini, A.; Tucci, P.; Celardo, I.; Melino, G. Senescence and aging: The critical roles of p53. Oncogene 2013, 32, 5129–5143. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.B.; Schumacher, B. p53 in the DNA-damage-repair process. Cold Spring Harb. Perspect. Med. 2016, 6, a026070. [Google Scholar] [CrossRef]
- Vogelstein, B.; Lane, D.; Levine, A.J. Surfing the p53 network. Nature (London, U.K.) 2000, 408, 307–310. [Google Scholar] [CrossRef] [PubMed]
- Hainaut, P.; Hollstein, M. p53 and human cancer: The first ten thousand mutations. Adv. Cancer Res. 1999, 77, 81–137. [Google Scholar]
- Wang, X. p53 regulation: Teamwork between RING domains of MDM2 and MDMX. Cell Cycle 2011, 10, 4225–4229. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Bayle, J.H.; Olson, D.; Levine, A.J. The p53-MDM2 autoregulatory feedback loop. Genes Dev. 1993, 7, 1126–1132. [Google Scholar] [CrossRef] [PubMed]
- Momand, J.; Wu, H.H.; Dasgupta, G. MDM2 - master regulator of the p53 tumor suppressor protein. Gene 2000, 242, 15–29. [Google Scholar] [CrossRef]
- Momand, J.; Zambetti, G.P.; Olson, D.C.; George, D.; Levine, A.J. The MDM2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 1992, 69, 1237–1245. [Google Scholar] [CrossRef]
- Oliner, J.D.; Pietenpol, J.A.; Thiagalingam, S.; Gyuris, J.; Kinzler, K.W.; Vogelstein, B. Oncoprotein MDM2 conceals the activation domain of tumor supressor p53. Nature (London, U.K.) 1993, 362, 857–860. [Google Scholar] [CrossRef]
- Roth, J.; Dobbelstein, M.; Freedman, D.A.; Shenk, T.; Levine, A.J. Nucleo-cytoplasmic shuttling of the HDM2 oncoprotein regulated the levels of the p53 protein via a pathway used by the human immunodeficiency vírus ver protein. EMBO J. 1998, 17, 554–564. [Google Scholar] [CrossRef]
- Haupt, Y.; Maya, R.; Kazaz, A.; Oren, M. MDM2 promotes the rapid degradation of p53. Nature 1997, 387, 296–299. [Google Scholar] [CrossRef] [PubMed]
- Pant, V.; Lozano, G. Limiting the power of p53 through the ubiquitin proteasome pathway. Genes Dev. 2014, 28, 1739–1751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Aguilar, A.; Bernard, D.; Wang, S. Small-molecule inhibitors of the MDM2-p53 protein-protein interaction (MDM2 inhibitors) in clinical trials for cancer treatment: Miniperspective. J. Med. Chem. 2014, 58, 1038–1052. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhao, Y.; Aguilar, A.; Bernard, D.; Yang, C.Y. Targeting the MDM2-p53 protein-protein interaction for new cancer therapy: Progress and challenges. Cold Spring Harb. Perspect. Med. 2017, 7, a026245. [Google Scholar] [CrossRef]
- Vassilev, L.T.; Vu, B.T.; Graves, B.; Carvajal, D.; Podlaski, F.; Filipovic, Z.; Kong, N.; Kammlott, U.; Lukacs, C.; Klein, C.; Fotouhi, N.; Liu, E.A. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 2004, 303, 844–848. [Google Scholar] [CrossRef]
- Lemos, A.; Leão, M.; Soares, J.; Palmeira, A.; Pinto, M.; Saraiva, L.; Sousa, M.E. Medicinal chemistry strategies to disrupt p53-MDM2/MDMX interaction. Med. Res. Rev. 2016, 36, 789–844. [Google Scholar] [CrossRef]
- Tisato, V.; Voltan, R.; Gonelli, A.; Secchiero, P.; Zauli, G. MDM2/X inhibitors under clinical evaluation: Perspectives for the management of hematological malignancies and pediatric cancer. J. Hematol. Oncol. 2017, 10, 133. [Google Scholar] [CrossRef] [PubMed]
- Pinto, M.M.M.; Sousa, M.E.; Nascimento, M.S. Xanthone derivatives: New insights in biological activities. Curr. Med. Chem. 2005, 12, 2517–2538. [Google Scholar] [CrossRef] [PubMed]
- Pouli, N.; Marakos, P. Fused xanthone derivatives as antiproliferative agents. Anticancer Agents Med. Chem. 2009, 9, 77–98. [Google Scholar] [CrossRef]
- Pedro, M.; Cerqueira, F.; Sousa, M.E.; Nascimento, M.S.J.; Pinto, M. Xanthones as inhibitors of growth of human cancer cell lines and their effects on the proliferation of human lymphocytes in vitro. Bioorg. Med. Chem. 2002, 10, 3725–3730. [Google Scholar] [CrossRef]
- Sousa, E.P.; Silva, A.M.S.; Pinto, M.M.M.; Pedro, M.M.; Cerqueira, F.A.M.; Nascimento, M.S.J. Isomeric kielcorins and dihydroxyxanthones: Synthesis, structure elucidation, and inhibitory activities of growth of human cancer cell lines and on the proliferation of human lymphocytes in vitro. Helv. Chim. Acta 2002, 85, 2862–2876. [Google Scholar] [CrossRef]
- Castanheiro, R.A.; Pinto, M.M.; Silva, A.M.; Cravo, S.M.; Gales, L.; Damas, A.M.; Nazareth, N.; Nascimento, M.S.J.; Eaton, G. Dihydroxyxanthones prenylated derivatives: Synthesis, structure elucidation, and growth inhibitory activity on human tumor cell lines with improvement of selectivity for MCF-7. Bioorg. Med. Chem. 2007, 15, 6080–6088. [Google Scholar] [CrossRef]
- Sousa, E.; Paiva, A.; Nazareth, N.; Gales, L.; Damas, A.M.; Nascimento, M.S.J.; Pinto, M. Bromoalkoxyxanthones as promising antitumor agents: Synthesis, crystal structure and effect on human tumor cell lines. Eur. J. Med. Chem. 2009, 44, 3830–3835. [Google Scholar] [CrossRef]
- Palmeira, A.; Paiva, A.; Sousa, E.; Seca, H.; Almeida, G.M.; Lima, R.T.; Fernandes, M.X.; Pinto, M.; Vasconcelos, M.H. Insights into the in vitro antitumor mechanism of action of a new pyranoxanthone. Chem. Biol. Drug Des. 2010, 76, 43–58. [Google Scholar] [CrossRef]
- Paiva, A.M.; Sousa, M.E.; Camoes, A.; Nascimento, M.S.J.; Pinto, M.M.M. Prenylated xanthones: Antiproliferative effects and enhancement of the growth inhibitory action of 4-hydroxytamoxifen in estrogen receptor-positive breast cancer cell line. Med. Chem. Res. 2012, 21, 552–558. [Google Scholar] [CrossRef]
- Leão, M.; Gomes, S.; Pedraza-Chaverri, J.; Machado, N.; Sousa, E.; Pinto, M.; Inga, A.; Pereira, C.; Saraiva, L. α-Mangostin and gambogic acid as potential inhibitors of the p53-MDM2 interaction revealed by a yeast-based approach. J. Nat. Prod. 2013, 76, 774–778. [Google Scholar] [CrossRef]
- Leão, M.; Pereira, C.; Bisio, A.; Ciribilli, Y.; Paiva, A.M.; Machado, N.; Palmeira, A.; Fernandes, M.X.; Sousa, E.; Pinto, M.; Inga, A.; Saraiva, L. Discovery of a new small-molecule inhibitor of p53-MDM2 interaction using a yeast-based approach. Biochem. Pharmacol. 2013, 85, 1234–1245. [Google Scholar] [CrossRef]
- Fernandes, C.; Masawang, K.; Tiritan, M.E.; Sousa, E.; de Lima, V.; Afonso, C.; Bousbaa, H.; Sudprasert, W.; Pedro, M.; Pinto, M. New chiral derivatives of xanthones: Synthesis and investigation of enantioselectivity as inhibitors of growth of human tumor cell lines. Bioorg. Med. Chem. 2014, 22, 1049–1062. [Google Scholar] [CrossRef]
- Barbosa, J.; Lima, R.T.; Sousa, D.; Gomes, A.S.; Palmeira, A.; Seca, H.; Choosang, K.; Pakkong, P.; Bousbaa, H.; Pinto, M.M.; et al. Screening a small library of xanthones for antitumor activity and identification of a hit compound which induces apoptosis. Molecules 2016, 21, 81. [Google Scholar] [CrossRef]
- Cidade, H.; Rocha, V.; Palmeira, A.; Marques, C.; Tiritan, M.E.; Ferreira, H.; Lobo, J.S.; Almeida, I.F.; Sousa, M.E.; Pinto, M. In silico and in vitro antioxidant and cytotoxicity evaluation of oxygenated xanthone derivatives. Arab. J. Chem. 2017. [Google Scholar] [CrossRef]
- Alves, A.; Correia-da-Silva, M.; Nunes, C.; Campos, J.; Sousa, E.; Silva, P.M.A.; Bousbaa, H.; Rodrigues, F.; Ferreira, D.; Costa, P.C.; et al. Discovery of a new xanthone against glioma: Synthesis and development of (pro)liposome formulations. Molecules 2019, 24, 409. [Google Scholar] [CrossRef]
- Gomes, S.; Raimundo, L.; Soares, J.; Loureiro, J.B.; Leão, M.; Ramos, H.; Monteiro, M.N.; Lemos, A.; Moreira, J.; Pinto, M.; et al. New inhibitor of the TAp73 interaction with MDM2 and mutant p53 with promising antitumor activity against neuroblastoma. Cancer Lett. 2019, 446, 90–102. [Google Scholar] [CrossRef]
- Liu, J.; Zhou, F.; Zhang, L.; Wang, H.; Zhang, J.; Zhang, C.; Jiang, Z.; Li, Y.; Liu, Z.; Chen, H. DMXAA-pyranoxanthone hybrids enhance inhibition activities against human cancer cells with multi-target functions. Eur. J. Med. Chem. 2018, 143, 1768–1778. [Google Scholar] [CrossRef]
- Hardcastle, I.R.; Liu, J.; Valeur, E.; Watson, A.; Ahmed, S.U.; Blackburn, T.J.; Bennaceur, K.; Clegg, W.; Drummond, C.; Endicott, J.A.; et al. Isoindolinone inhibitors of the murine double minute 2 (MDM2)-p53 protein-protein interaction: Structure-activity studies leading to improved potency. J. Med. Chem. 2011, 54, 1233–1243. [Google Scholar] [CrossRef]
- Twarda-Ciapa, A.; Krzanik, S.; Kubica, K.; Guzik, K.; Labuzek, B.; Neochoritis, C.G.; Khoury, K.; Kowalska, K.; Czub, M.; Dubin, G.; et al. 1,4,5-Trisubstituted imidazole-based p53-MDM2/MDMX antagonists with aliphatic linkers for conjugation with biological carriers. J. Med. Chem. 2017, 60, 4234–4244. [Google Scholar] [CrossRef]
- Gicquel, M.; Gomez, C.; Alvarez, M.C.G.; Pamlard, O.; Guérineau, V.; Jacquet, E.; Bignon, J.; Voituriez, A.; Marinetti, A. Inhibition of p53-murine double minute 2 (MDM2) interactions with 3,3′-spirocyclopentene oxindole derivatives. J. Med. Chem. 2018, 61, 9386–9392. [Google Scholar] [CrossRef]
- Quillinan, A.J.; Schelnmann, F. Studies in the xanthone series. Part XII. A general synthesis of polyoxygenated xanthones from benzophenone precursors. J. Chem. Soc. Perkin Trans. 1973, 1, 1329–1337. [Google Scholar] [CrossRef]
- Resende, D.I.S.P.; Pereira-Terra, P.; Inácio, Â.S.; da Costa, P.M.; Pinto, E.; Sousa, E.; Pinto, M.M.M. Lichen xanthones as models for new antifungal agents. Molecules 2018, 23, 2617. [Google Scholar] [CrossRef]
- Martins, E.; Silva, V.; Lemos, A.; Palmeira, A.; Puthongking, P.; Sousa, E.; Rocha-Pereira, C.; Ghanem, C.I.; Carmo, H.; Remião, F.; et al. Newly synthesized oxygenated xanthones as potential P-glycoprotein activators: In vitro, ex vivo, and in silico studies. Molecules 2019, 24, 707. [Google Scholar] [CrossRef]
- Soares, J.; Espadinha, M.; Raimundo, L.; Ramos, H.; Gomes, A.S.; Gomes, S.; Loureiro, J.B.; Inga, A.; Reis, F.; Gomes, C.; et al. DIMP53-1: A novel small-molecule dual inhibitor of p53-MDM2/X interactions with multifunctional p53-dependent anticancer properties. Mol. Oncol. 2017, 11, 612–627. [Google Scholar] [CrossRef]
- Soares, J.; Pereira, N.A.; Monteiro, Â.; Leão, M.; Bessa, C.; dos Santos, D.J.; Raimundo, L.; Queiroz, G.; Bisio, A.; Inga, A.; et al. Oxazoloisoindolinones with in vitro antitumor activity selectively activate a p53-pathway through potential inhibition of the p53-MDM2 interaction. Eur. J. Pharm. Sci. 2015, 66, 138–147. [Google Scholar] [CrossRef]
- Soares, J.; Raimundo, L.; Pereira, N.A.; dos Santos, D.J.; Pérez, M.; Queiroz, G.; Leão, M.; Santos, M.M.M.; Saraiva, L. A tryptophanol-derived oxazolopiperidone lactam is cytotoxic against tumors via inhibition of p53 interaction with murine double minute proteins. Pharmacol. Res. 2015, 95–96, 42–52. [Google Scholar] [CrossRef]
- Chen, L.; Yin, H.; Farooqi, B.; Sebti, S.; Hamilton, A.D.; Chen, J. p53 α-helix mimetics antagonize p53/MDM2 interaction and activate p53. Mol. Cancer Ther. 2005, 4, 1019–1025. [Google Scholar] [CrossRef]
- Kalid, O.; Ben-Tal, N. Study of MDM2 binding to p53-analogues: Affinity, helicity, and applicability to drug design. J. Chem. Inf. Model. 2009, 49, 865–876. [Google Scholar] [CrossRef]
- Kussie, P.H.; Gorina, S.; Marechal, V.; Elenbaas, B.; Moreau, J.; Levine, A.J.; Pavletich, N.P. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science 1996, 274, 948–953. [Google Scholar] [CrossRef]
- Anil, B.; Riedinger, C.; Endicott, J.A.; Nobie, M.E. The structure of an MDM2-nutlin-3a complex solved by the use of a validated MDM2 surface-entropy reduction mutant. Acta Crystallogr. D Biol. Crystallogr. 2013, 69, 1358–1366. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and the accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Bharatham, N.; Bharatham, K.; Shelat, A.A.; Bashford, D. Ligand binding mode prediction by docking: MDM2/MDMX inhibitors as a case study. J. Chem. Inf. Model. 2014, 54, 648–659. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xiao, J.; Sujek, T.O.; Zhang, J.; Wang, J.; Bryant, S.H. PubChem: A public information system for analyzing bioactivities of small molecules. Nucleic Acids Res. 2009, 37, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Kini, R.M.; Evans, H.J. Molecular modeling of proteins: A strategy for energy minimization by molecular mechanics in the AMBER force field. J. Biomol. Struct. Dyn. 1991, 9, 475–488. [Google Scholar] [CrossRef] [PubMed]
- Seeliger, D.; de Groot, B.L. Ligand docking and binding site analysis with PyMOL and AutoDock/Vina. J. Comput. Aided Mol. Des. 2010, 24, 417–422. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds 5, 16–40 are available from the authors. |








| Xanthones 30–40 | |
| Ligands | Free Energy of the Ligand: MDM2 (kcal·mol−1) |
| 30 | −6.6 |
| 31 | −6.2 |
| 32 | −6.6 |
| 33 | −5.9 |
| 34 | −6.9 |
| 35 | −7.2 |
| 36 | −6.6 |
| 37 | −7.3 |
| 38 | −7.1 |
| 39 | −7.3 |
| 40 | −7.4 |
| Inhibitors of MDM2-p53 Interaction | |
| Ligands | Free Energy of the Ligand: MDM2 (kcal·mol−1) |
| 1 | −7.7 |
| 2 | −8.0 |
| 3 | −7.9 |
| 4 | −8.0 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lemos, A.; Gomes, A.S.; Loureiro, J.B.; Brandão, P.; Palmeira, A.; Pinto, M.M.M.; Saraiva, L.; Sousa, M.E. Synthesis, Biological Evaluation, and In Silico Studies of Novel Aminated Xanthones as Potential p53-Activating Agents. Molecules 2019, 24, 1975. https://doi.org/10.3390/molecules24101975
Lemos A, Gomes AS, Loureiro JB, Brandão P, Palmeira A, Pinto MMM, Saraiva L, Sousa ME. Synthesis, Biological Evaluation, and In Silico Studies of Novel Aminated Xanthones as Potential p53-Activating Agents. Molecules. 2019; 24(10):1975. https://doi.org/10.3390/molecules24101975
Chicago/Turabian StyleLemos, Agostinho, Ana Sara Gomes, Joana B. Loureiro, Pedro Brandão, Andreia Palmeira, Madalena M. M. Pinto, Lucília Saraiva, and Maria Emília Sousa. 2019. "Synthesis, Biological Evaluation, and In Silico Studies of Novel Aminated Xanthones as Potential p53-Activating Agents" Molecules 24, no. 10: 1975. https://doi.org/10.3390/molecules24101975