Technologies for the Selection, Culture and Metabolic Profiling of Unique Rhizosphere Microorganisms for Natural Product Discovery
Abstract
:1. Introduction
2. Results
2.1. Phylogenetic Marker Gene Sequencing for Identification of Bacterial and Fungal Taxa in the Canola Rhizosphere
2.2. Identification of Selected Bacterial Colonies by Sanger Sequencing of 16S rRNA
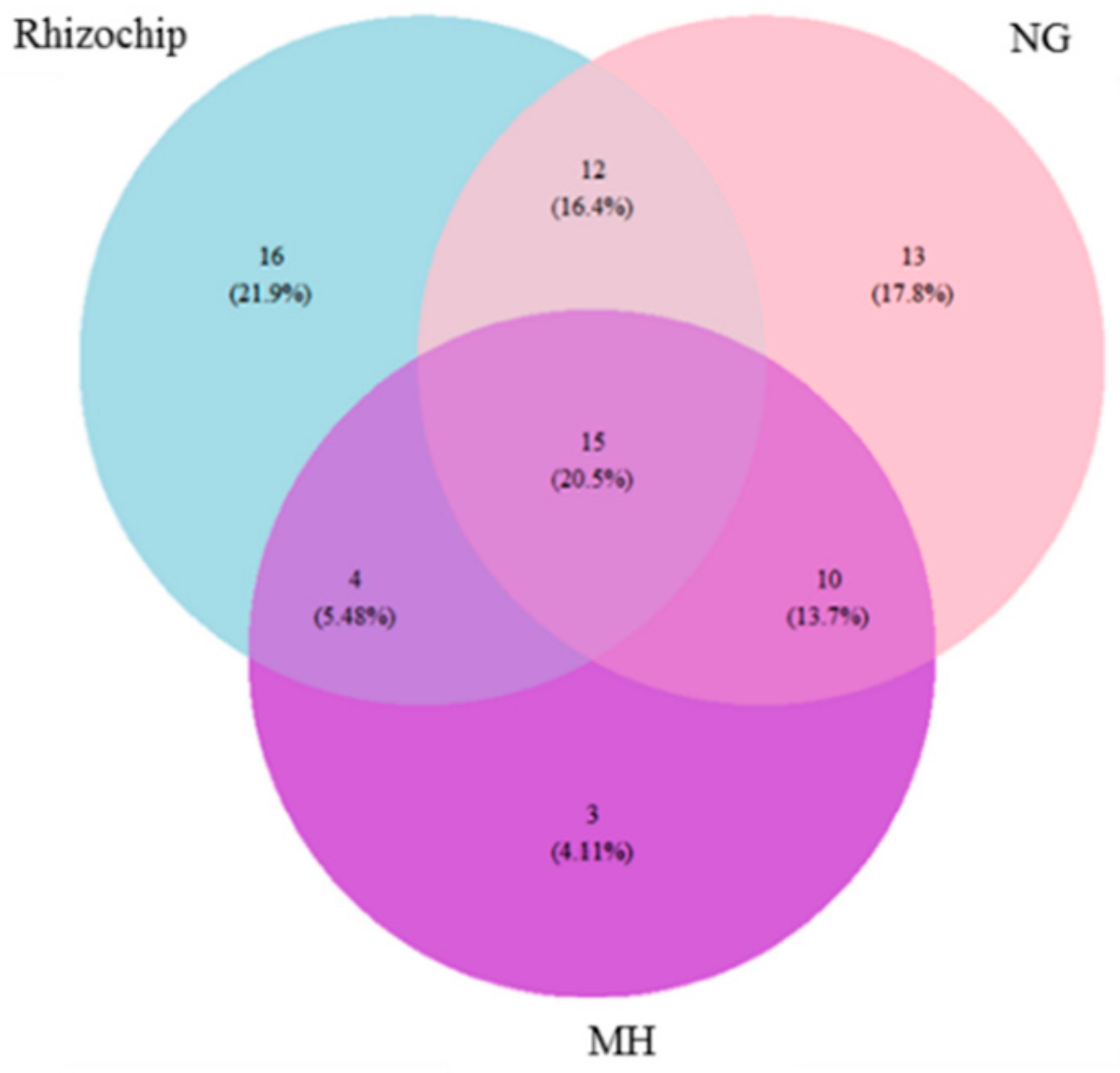
2.3. Genomic Identification of Isolated Microbial Cohorts Through Standard- and Rhizochip-Based Culture Methods
2.4. Identification of Selected Bacterial Colonies by Sanger Sequencing of 16S rRNA
2.5. Metabolic Profiling of Selected Organisms
3. Discussion
4. Materials and Methods
4.1. Establishment of the Canola Field Trial
4.2. Soil Sampling
4.3. In Vitro Culture of Soil Microorganisms Obtained through Soil Suspensions in Various Culture Media and Solidifying Agents
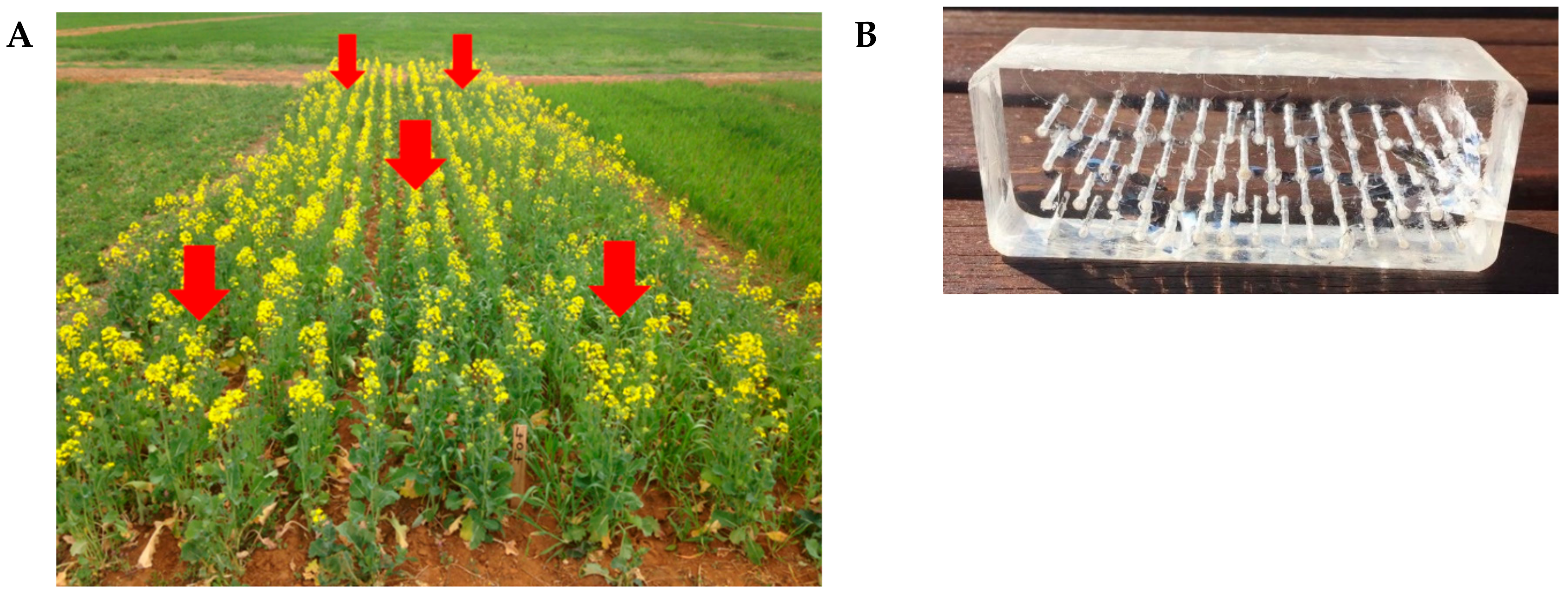
4.4. In Situ Culture with a Prototype Rhizochip Apparatus and Isolation of Microbial Colonies
4.5. Genomic DNA Extraction from Isolated Microbial Colonies and Phylogenetic Marker Gene Sequencing Targeting Bacterial 16S rRNA and Fungal ITS Regions
4.6. Genomic DNA Extraction from Rhizosphere Soils
4.7. Extraction of Soil Microbial Metabolites and Metabolic Profiling
4.8. Statistical Analysis
4.8.1. Enumeration and Characterisation of Soil Microbial Cultures
4.8.2. Metagenomics Analysis
4.8.3. Metabolic Profiling
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dennis, P.G.; Miller, A.J.; Hirsch, P.R. Are root exudates more important than other sources of rhizodeposits in structuring rhizosphere bacterial communities? FEMS Microbiol. Ecol. 2010, 72, 313–327. [Google Scholar] [Green Version]
- Weyens, N.; van der Lelie, D.; Taghavi, S.; Newman, L.; Vangronsveld, J. Exploiting plant-microbe partnerships to improve biomass production and remediation. Trends Biotechnol. 2009, 27, 591–598. [Google Scholar] [CrossRef] [PubMed]
- Berg, G. Plant-microbe interactions promoting plant growth and health: perspectives for controlled use of microorganisms in agriculture. Appl. Microbiol. Biotechnol. 2009, 84, 11–18. [Google Scholar] [CrossRef]
- Berendsen, R.L.; Pieterse, C.M.J.; Bakker, P.A.H.M. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Raaijmakers, J.M.; Mazzola, M. Diversity and natural functions of antibiotics produced by beneficial and plant pathogenic bacteria. Annu. Rev. Phytopathol. 2012, 50, 403–424. [Google Scholar] [CrossRef] [PubMed]
- Piddock, L.J. Teixobactin, the first of a new class of antibiotics discovered by ichip technology? J. Antimicrob. Chemother. 2015, 70, 2679–2680. [Google Scholar] [CrossRef]
- Sansinenea, E.; Ortiz, A. Secondary metabolites of soil Bacillus spp. Biotechnol. Lett. 2011, 33, 1523–1538. [Google Scholar] [CrossRef]
- Buckeridge, K.M.; Banerjee, S.; Siciliano, S.D.; Grogan, P. The seasonal pattern of soil microbial community structure in mesic low arctic tundra. Soil Biol. Biochem. 2013, 65, 338–347. [Google Scholar] [CrossRef]
- Dunfield, K.E.; Germida, J.J. Seasonal changes in the rhizosphere microbial communities associated with field-grown genetically modified canola (Brassica napus). Appl. Environ. Microbiol. 2003, 69, 7310–7318. [Google Scholar] [CrossRef]
- Lam, K.S. New aspects of natural products in drug discovery. Trends Microbiol. 2007, 15, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Banu, N.A.; Singh, B.; Copeland, L. Microbial biomass and microbial biodiversity in some soils from New South Wales, Australia. Soil Res. 2004, 42, 777–782. [Google Scholar] [CrossRef]
- Edwards, J.; Johnson, C.; Santos-Medellín, C.; Lurie, E.; Podishetty, N.K.; Bhatnagar, S.; Eisen, J.A.; Sundaresan, V. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. USA 2015, 112, E911–E920. [Google Scholar] [CrossRef] [Green Version]
- Singh, B.K.; Millard, P.; Whiteley, A.S.; Murrell, J.C. Unravelling rhizosphere--microbial interactions: opportunities and limitations. Trends Microbiol. 2004, 12, 386–393. [Google Scholar] [CrossRef] [PubMed]
- Torsvik, V.; Øvreås, L. Microbial diversity and function in soil: From genes to ecosystems. Curr. Opin. Microbiol. 2002, 5, 240–245. [Google Scholar] [CrossRef]
- Ling, L.L.; Schneider, T.; Peoples, A.J.; Spoering, A.L.; Engels, I.; Conlon, B.P.; Mueller, A.; Schaberle, T.F.; Hughes, D.E.; Epstein, S.; et al. A new antibiotic kills pathogens without detectable resistance. Nature 2015, 517, 455–459. [Google Scholar] [CrossRef] [PubMed]
- Sørensen, J. The Rhizosphere as a Habitat for Soil Microorganisms. In Modern Soil Microbiology; van Elsas, J.D., Trevors, J.T., Wellington, E.M.H., Eds.; Marcel Dekker Inc.: New York, NY, USA, 1997; pp. 21–45. [Google Scholar]
- Janssen, P.H. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 2006, 72, 1719 LP–1728 LP. [Google Scholar] [CrossRef] [PubMed]
- Kaewkla, O.; Franco, C.M.M. Rational approaches to improving the isolation of endophytic actinobacteria from Australian native trees. Microb. Ecol. 2013, 65, 384–393. [Google Scholar] [CrossRef] [PubMed]
- Traxler, M.F.; Kolter, R. A massively spectacular view of the chemical lives of microbes. Proc. Natl. Acad. Sci. USA 2012, 109, 10128–10129. [Google Scholar] [CrossRef] [Green Version]
- Vieira, F.C.S.; Nahas, E. Comparison of microbial numbers in soils by using various culture media and temperatures. Microbiol. Res. 2005, 160, 197–202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis, K.E.R.; Sangwan, P.; Janssen, P.H. Acidobacteria, Rubrobacteridae and Chloroflexi are abundant among very slow-growing and mini-colony-forming soil bacteria. Environ. Microbiol. 2011, 13, 798–805. [Google Scholar] [CrossRef]
- Nichols, D.; Cahoon, N.; Trakhtenberg, E.M.; Pham, L.; Mehta, A.; Belanger, A.; Kanigan, T.; Lewis, K.; Epstein, S.S. Use of ichip for high-throughput in situ cultivation of “uncultivable” microbial species. Appl. Environ. Microbiol. 2010, 76, 2445–2450. [Google Scholar] [CrossRef] [PubMed]
- Jung, D.; Aoi, Y.; Epstein, S.S. In situ cultivation allows for recovery of bacterial types competitive in their natural environment. Microbes Environ. 2016, 31, 456–459. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N.; Leff, J.W.; Adams, B.J.; Nielsen, U.N.; Bates, S.T.; Lauber, C.L.; Owens, S.; Gilbert, J.A.; Wall, D.H.; Caporaso, J.G. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc. Natl. Acad. Sci. USA 2012, 109, 21390–21395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weston, L.; Skoneczny, D.; Weston, P.; Weidenhamer, J.D. Metabolic profiling: An overview—New approaches for the detection and functional analysis of biologically active secondary plant products. J. Allelochem. Interact. 2015, 1, 15–27. [Google Scholar]
- Bertrand, S.; Bohni, N.; Schnee, S.; Schumpp, O.; Gindro, K.; Wolfender, J.L. Metabolite induction via microorganism co-culture: A potential way to enhance chemical diversity for drug discovery. Biotechnol. Adv. 2014, 32, 1180–1204. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.-L.; Xu, Y.; Straight, P.D.; Dorrestein, P.C. Translating metabolic exchange with imaging mass spectrometry. Nat. Chem. Biol. 2009, 5, 885–887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watrous, J.; Roach, P.; Alexandrov, T.; Heath, B.S.; Yang, J.Y.; Kersten, R.D.; van der Voort, M.; Pogliano, K.; Gross, H.; Raaijmakers, J.M.; et al. Mass spectral molecular networking of living microbial colonies. Proc. Natl. Acad. Sci. USA 2012, 109, E1743–E1752. [Google Scholar] [CrossRef] [Green Version]
- Traxler, M.F.; Watrous, J.D.; Alexandrov, T.; Dorrestein, P.C.; Kolter, R. Interspecies interactions stimulate diversification of the Streptomyces coelicolor secreted metabolome. MBio 2013, 4, e00459-13. [Google Scholar] [CrossRef]
- Weston, L.A.; Mathesius, U. Flavonoids: Their structure, biosynthesis and role in the rhizosphere, including allelopathy. J. Chem. Ecol. 2013, 39, 283–297. [Google Scholar] [CrossRef]
- Pérez-Jaramillo, J.E.; Mendes, R.; Raaijmakers, J.M. Impact of plant domestication on rhizosphere microbiome assembly and functions. Plant Mol. Biol. 2016, 90, 635–644. [Google Scholar] [CrossRef]
- Bertin, C.; Yang, X.; Weston, L.A. The role of root exudates and allelochemicals in the rhizosphere. Plant Soil 2003, 256, 67–83. [Google Scholar] [CrossRef]
- Cox, D.G.; Oh, J.; Keasling, A.; Colson, K.L.; Hamann, M.T. The utility of metabolomics in natural product and biomarker characterization. Biochim. Biophys. Acta 2014, 1840, 3460–3474. [Google Scholar] [CrossRef] [Green Version]
- Rosato, A.; Tenori, L.; Cascante, M.; De Atauri Carulla, P.R.; Martins Dos Santos, V.A.P.; Saccenti, E. From correlation to causation: analysis of metabolomics data using systems biology approaches. Metabolomics 2018, 14, 37. [Google Scholar] [CrossRef] [PubMed]
- Bissett, A.; Fitzgerald, A.; Meintjes, T.; Mele, P.M.; Reith, F.; Dennis, P.G.; Breed, M.F.; Brown, B.; Brown, M.V.; Brugger, J.; et al. Introducing BASE: The Biomes of Australian Soil Environments soil microbial diversity database. Gigascience 2016, 5, 21. [Google Scholar] [CrossRef] [PubMed]
- Dingman, D.W.; Stahly, D.P. Medium promoting sporulation of Bacillus larvae and metabolism of medium components. Appl. Environ. Microbiol. 1983, 46, 860–869. [Google Scholar]
- Munsch, P.; Alatossava, T.; Marttinen, N.; Meyer, J.-M.; Christen, R.; Gardan, L. Pseudomonas costantinii sp. nov., another causal agent of brown blotch disease, isolated from cultivated mushroom sporophores in Finland. Int. J. Syst. Evol. Microbiol. 2002, 52, 1973–1983. [Google Scholar] [PubMed]
- Zhao, L.; Yang, X.; Li, X.; Wei, M.U.; Feng, L.I.U. Antifungal, insecticidal and herbicidal properties of volatile components from Paenibacillus polymyxa strain BMP-11. Agric. Sci. China 2011, 10, 728–736. [Google Scholar] [CrossRef]
- Kotoučková, L.; Schumann, P.; Durnova, E.; Spröer, C.; Sedláček, I.; Neča, J.; Zdráhal, Z.; Němec, M. Arthrobacter nitroguajacolicus sp. nov., a novel 4-nitroguaiacol-degrading actinobacterium. Int. J. Syst. Evol. Microbiol. 2004, 54, 773–777. [Google Scholar] [CrossRef]
- Abdel-El-Haleem, D. Acinetobacter: Environmental and biotechnological applications. African J. Biotechnol. 2003, 2, 71–74. [Google Scholar]
- De Oliveira, E.J.; Rabinovitch, L.; Monnerat, R.G.; Passos, L.K.J.; Zahner, V. Molecular characterization of Brevibacillus laterosporus and its potential use in biological control. Appl. Environ. Microbiol. 2004, 70, 6657–6664. [Google Scholar] [CrossRef] [PubMed]
- Han, J.I.; Choi, H.K.; Lee, S.W.; Orwin, P.M.; Kim, J.; LaRoe, S.L.; Kim, T.G.; O’Neil, J.; Leadbetter, J.R.; Lee, S.Y.; et al. Complete genome sequence of the metabolically versatile plant growth-promoting endophyte Variovorax paradoxus S110. J. Bacteriol. 2011, 193, 1183–1190. [Google Scholar] [CrossRef] [PubMed]
- Chandra, D.; Srivastava, R.; Gupta, V.V.S.R.; Franco, C.M.M.; Sharma, A.K. Evaluation of ACC-deaminase-producing rhizobacteria to alleviate water-stress impacts in wheat (Triticum aestivum L.) plants. Can. J. Microbiol. 2019, 65, 1–17. [Google Scholar] [CrossRef]
- Belimov, A.A.; Dodd, I.C.; Hontzeas, N.; Theobald, J.C.; Safronova, V.I.; Davies, W.J. Rhizosphere bacteria containing 1-aminocyclopropane-1-carboxylate deaminase increase yield of plants grown in drying soil via both local and systemic hormone signalling. New Phytol. 2009, 181, 413–423. [Google Scholar] [CrossRef] [PubMed]
- Satola, B.; Wübbeler, J.H.; Steinbüchel, A. Metabolic characteristics of the species Variovorax paradoxus. Appl. Microbiol. Biotechnol. 2013, 97, 541–560. [Google Scholar] [CrossRef]
- Aislabie, J.; Bej, A.K.; Ryburn, J.; Lloyd, N.; Wilkins, A. Characterization of Arthrobacter nicotinovorans HIM, an atrazine-degrading bacterium, from agricultural soil New Zealand. FEMS Microbiol. Ecol. 2005, 52, 279–286. [Google Scholar] [CrossRef]
- Rohr, J.; Eick, S.; Zeeck, A.; Reuschenbach, P.; Zahner, H.; Fiedler, H.-P. Metabolic products of microorganisms. 249 Tetracenomycins B3 and D3, key intermediates of the elloramycin and tetracenomycin C biosynthesis. J. Antibiot. 1988, 8, 1066–1073. [Google Scholar] [CrossRef]
- MetaCyc Pathway: Tetracenomycin C biosynthesis. Available online: https://biocyc.org/META/NEW-IMAGE?type=PATHWAY&object=PWY-7485. (accessed on 17 May 2019).
- Sasaki, T.; Li, W.; Zaike, S.; Asada, Y.; Li, Q.; Ma, F.; Zhang, Q.; Koike, K. Antioxidant lignoids from leaves of Ribes nigrum. Phytochemistry 2013, 95, 333–340. [Google Scholar] [CrossRef]
- Hongming, L.; Xu, L.; Zhaojian, G.; Fan, Y.; Dingbin, C.; Jianchun, Z.; Jianhong, X.; Shunpeng, L.; Qing, H. Isolation of an aryloxyphenoxy propanoate (AOPP) herbicide-degrading strain Rhodococcus ruber JPL-2 and the cloning of a novel carboxylesterase gene (feh). Braz. J. Microbiol. 2015, 46, 425–432. [Google Scholar] [CrossRef]
- Katz, M.; Hover, B.M.; Brady, S.F. Culture-independent discovery of natural products from soil metagenomes. J. Ind. Microbiol. Biotechnol. 2016, 43, 129–141. [Google Scholar] [CrossRef]
- Fleming, A. On the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. influenzae. Br. J. Exp. Pathol. 1929, 10, 226. [Google Scholar] [CrossRef]
- Pickett, J.A.; Weston, L.A. Possibilities for rationally exploiting co-evolution in addressing resistance to insecticides, and beyond. Pestic. Biochem. Physiol. 2018, 151, 18–24. [Google Scholar] [CrossRef]
- Shade, A.; Hogan, C.S.; Klimowicz, A.K.; Linske, M.; McManus, P.S.; Handelsman, J. Culturing captures members of the soil rare biosphere. Environ. Microbiol. 2012, 14, 2247–2252. [Google Scholar] [CrossRef]
- Nazir, R.; Warmink, J.A.; Boersma, H.; Elsas, J.D. Mechanisms that promote bacterial fitness in fungal-affected soil microhabitats. FEMS Microbiol. Ecol. 2010, 71. [Google Scholar] [CrossRef]
- Bainard, L.D.; Hamel, C.; Gan, Y. Edaphic properties override the influence of crops on the composition of the soil bacterial community in a semiarid agroecosystem. Appl. Soil Ecol. 2016, 105, 160–168. [Google Scholar] [CrossRef]
- Zorner, P.; Farmer, S.; Alibek, K. Quantifying crop rhizosphere microbiome ecology: The next frontier in enhancing the commercial utility of agricultural microbes. Ind. Biotechnol. 2018, 14, 116–119. [Google Scholar] [CrossRef]
- De Coninck, B.; Timmermans, P.; Vos, C.; Cammue, B.P.A.; Kazan, K. What lies beneath: Belowground defense strategies in plants. Trends Plant Sci. 2015, 20, 91–101. [Google Scholar] [CrossRef]
- Janssen, P.H.; Yates, P.S.; Grinton, B.E.; Taylor, P.M.; Sait, M.; Janssen, P.H.; Yates, P.S.; Grinton, B.E.; Taylor, P.M.; Sait, M. Improved Culturability of Soil Bacteria and Isolation in Pure Culture of Novel Members of the Divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl. Environ. Microbiol. 2002, 68, 2391–2396. [Google Scholar] [CrossRef]
- Lay, C.-Y.; Bell, T.H.; Hamel, C.; Harker, K.N.; Mohr, R.; Greer, C.W.; Yergeau, É.; St-Arnaud, M. Canola Root--Associated Microbiomes in the Canadian Prairies. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef]
- Chaparro, J.M.; Badri, D.V.; Vivanco, J.M. Rhizosphere microbiome assemblage is affected by plant development. ISME J. 2014, 8, 790. [Google Scholar] [CrossRef]
- Broeckling, C.D.; Broz, A.K.; Bergelson, J.; Manter, D.K.; Vivanco, J.M. Root exudates regulate soil fungal community composition and diversity. Appl. Environ. Microbiol. 2008, 74, 738–744. [Google Scholar] [CrossRef]
- Buyer, J.S.; Teasdale, J.R.; Roberts, D.P.; Zasada, I.A.; Maul, J.E. Factors affecting soil microbial community structure in tomato cropping systems. Soil Biol. Biochem. 2010, 42, 831–841. [Google Scholar] [CrossRef]
- Buyer, J.S.; Baligar, V.C.; He, Z.; Arévalo-Gardini, E. Soil microbial communities under cacao agroforestry and cover crop systems in Peru. Appl. Soil Ecol. 2017, 120, 273–280. [Google Scholar] [CrossRef]
- Freedman, Z.; Zak, D.R. Soil bacterial communities are shaped by temporal and environmental filtering: evidence from a long-term chronosequence. Environ. Microbiol. 2015, 17, 3208–3218. [Google Scholar] [CrossRef]
- Sengupta, A.; Dick, W.A. Bacterial community diversity in soil under two tillage practices as determined by pyrosequencing. Microb. Ecol. 2015, 70, 853–859. [Google Scholar] [CrossRef]
- Marschner, P.; Yang, C.-H.; Lieberei, R.; Crowley, D.E. Soil and plant specific effects on bacterial community composition in the rhizosphere. Soil Biol. Biochem. 2001, 33, 1437–1445. [Google Scholar] [CrossRef]
- Mbuthia, L.W.; Acosta-Martinez, V.; DeBruyn, J.; Schaeffer, S.; Tyler, D.; Odoi, E.; Mpheshea, M.; Walker, F.; Eash, N. Long term tillage, cover crop, and fertilization effects on microbial community structure, activity: Implications for soil quality. Soil Biol. Biochem. 2015, 89, 24–34. [Google Scholar] [CrossRef]
- Blair, N.; Crocker, G.J. Crop rotation effects on soil carbon and physical fertility of two Australian soils. Soil Res. 2000, 38, 71–84. [Google Scholar] [CrossRef]
- Dalal, R.C.; Chan, K.Y. Soil organic matter in rainfed cropping systems of the Australian cereal belt. Soil Res. 2001, 39, 435–464. [Google Scholar] [CrossRef]
- Pham, V.H.T.; Kim, J. Cultivation of unculturable soil bacteria. Trends Biotechnol. 2012, 30, 475–484. [Google Scholar] [CrossRef]
- Kamagata, Y.; Tamaki, H. Cultivation of uncultured fastidious microbes. Microbes Environ. 2005, 20, 85–91. [Google Scholar] [CrossRef]
- Hamaki, T.; Suzuki, M.; Fudou, R.; Jojima, Y.; Kajiura, T.; Tabuchi, A.; Sen, K.; Shibai, H. Isolation of novel bacteria and actinomycetes using soil-extract agar medium. J. Biosci. Bioeng. 2005, 99, 485–492. [Google Scholar] [CrossRef]
- Kaeberlein, T.; Lewis, K.; Epstein, S.S. Isolating “uncultivatable” microorganisms in pure culture in a simulated natural environment. Science 2002, 296, 1127–1129. [Google Scholar] [CrossRef]
- Bhargava, N.; Sharma, P.; Capalash, N. Quorum sensing in Acinetobacter: an emerging pathogen. Crit. Rev. Microbiol. 2010, 36, 349–360. [Google Scholar] [CrossRef]
- Roca Subirà, I.; Espinal, P.; Vila-Farrés, X.; Vila Estapé, J. The Acinetobacter baumannii oxymoron: Commensal hospital dweller turned pan-drug-resistant menace. Front. Microbiol. 2012, 3, 148. [Google Scholar]
- Chapartegui-González, I.; Lázaro-D’iez, M.; Bravo, Z.; Navas, J.; Icardo, J.M.; Ramos-Vivas, J. Acinetobacter baumannii maintains its virulence after long-time starvation. PLoS ONE 2018, 13, e0201961. [Google Scholar] [CrossRef]
- Tetracenomycin C/8-demethyltetracenomycin C biosynthesis, tetracenomycin F2 => tetracenomycin C/8-demethyltetracenomycin C. Available online: https://www.genome.jp/kegg-bin/show_module?M00783 (accessed on 17 March 2019).
- Keikha, M. Williamsia spp. are emerging opportunistic bacteria. New Microbes New Infect. 2017, 21, 88–89. [Google Scholar] [CrossRef]
- Pathom-Aree, W.; Nogi, Y.; Sutcliffe, I.C.; Ward, A.C.; Horikoshi, K.; Bull, A.T.; Goodfellow, M. Williamsia marianensis sp. nov., a novel actinomycete isolated from the Mariana Trench. Int. J. Syst. Evol. Microbiol. 2006, 56, 1123–1126. [Google Scholar] [CrossRef]
- Horn, H.; Keller, A.; Hildebrandt, U.; Kämpfer, P.; Riederer, M.; Hentschel, U. Draft genome of the Arabidopsis thaliana phyllosphere bacterium, Williamsia sp. ARP1. Stand. Genomic Sci. 2016, 11, 8. [Google Scholar] [CrossRef]
- Teponno, R.B.; Kusari, S.; Spiteller, M. Recent advances in research on lignans and neolignans. Nat. Prod. Rep. 2016, 33, 1044–1092. [Google Scholar] [CrossRef] [Green Version]
- Fernández-Cabezón, L.; Galán, B.; Garcia, J.L. New insights on steroid biotechnology. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef]
- Yu, B.; Xu, P.; Shi, Q.; Ma, C. Deep desulfurization of diesel oil and crude oils by a newly isolated Rhodococcus erythropolis strain. Appl. Environ. Microbiol. 2006, 72, 54–58. [Google Scholar] [CrossRef]
- Mukram, I.; Ramesh, M.; Monisha, T.R.; Nayak, A.S.; Karegoudar, T.B. Biodegradation of butyronitrile and demonstration of its mineralization by Rhodococcus sp. MTB5. 3 Biotech. 2016, 6, 141. [Google Scholar] [CrossRef]
- McLeod, M.P.; Warren, R.L.; Hsiao, W.W.L.; Araki, N.; Myhre, M.; Fernandes, C.; Miyazawa, D.; Wong, W.; Lillquist, A.L.; Wang, D.; et al. The complete genome of Rhodococcus sp. RHA1 provides insights into a catabolic powerhouse. Proc. Natl. Acad. Sci. USA 2006, 103, 15582–15587. [Google Scholar] [CrossRef]
- Australian Genome Research Facility. Diversity Profiling Service Sample Preparation Guide. 2014. Available online: http://www.agrf.org.au/docs/diversity-profiling-sample-submission-guidelines.pdf (accessed on 24 November 2018).
- Turner, S.; Pryer, K.M.; Miao, V.P.W.; Palmer, J.D. Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis 1. J. Eukaryot. Microbiol. 1999, 46, 327–338. [Google Scholar] [CrossRef]
- Frykman, P.K.; Nordenskjöld, A.; Kawaguchi, A.; Hui, T.T.; Granström, A.L.; Cheng, Z.; Tang, J.; Underhill, D.M.; Iliev, I.; Funari, V.A.; et al. Characterization of bacterial and fungal microbiome in children with Hirschsprung disease with and without a history of enterocolitis: A multicenter study. PLoS ONE 2015, 10, e0124172. [Google Scholar] [CrossRef]
- Vogler, D.R.; Bruns, T.D. Phylogenetic relationships among the pine stem rust fungi (Cronartium and Peridermium spp.). Mycologia 1998, 90, 244–257. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996. [Google Scholar] [CrossRef]
- Kõljalg, U.; Larsson, K.-H.; Abarenkov, K.; Nilsson, R.H.; Alexander, I.J.; Eberhardt, U.; Erland, S.; Høiland, K.; Kjøller, R.; Larsson, E.; et al. UNITE: A database providing web-based methods for the molecular identification of ectomycorrhizal fungi. New Phytol. 2005, 166, 1063–1068. [Google Scholar] [CrossRef]
- Danoff-Burg, J.A.; Chen, X. Biodiversity Calculator 2005. Available online: http://www.columbia. edu/itc/cerc/danoffburg/Biodiversity\% 20Calculator. xls (accessed on 24 November 2018).
- Hammer, Ø.; Harper, D.A.T.; Ryan, P. PAST: Paleontological statistics software package for education and data analysis. Paleontol. Electron. 2001, 9. [Google Scholar]
- Chen, H.; Boutros, P.C. VennDiagram: A package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics 2011, 12, 35. [Google Scholar] [CrossRef]
- Team, R.C. R: A language and environment for statistical computing; R Foundation for Statistical Computing: Vienna, Austria; Available online: http://www.R-project.org. (accessed on 28 November 2005).
- Clarke, K.R. Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 1993, 18, 117–143. [Google Scholar] [CrossRef]
Sample Availability: Samples of extracts containing the compounds are available from the authors. |





| Colony ID | Source | Identification (Percent Similarity) | Agroecological Relevance | Reference |
|---|---|---|---|---|
| 24 | Rhizochip | Pseudomonas costantinii (100%) | Pathogen affecting mushrooms | [37] |
| 26 | Rhizochip | Pseudomonas sp. str. LaGso27l (98.7%) | ||
| 32 | Rhizochip | Paenibacillus polymyxa str. BMP-11 (100%) | Insecticidal and herbicidal activity | [38] |
| 37 | Rhizochip | Chryseobacterium sp. str. KR200 (100%) | ||
| 45 | Rhizochip | Chryseobacterium indologenes str. H2S10 (100%) | ||
| 238 | MH | Arthrobacter nitroguajacolicus str. G2-1 (100%) | 4-nitroguaiacol-degradation | [39] |
| 240 | MH | Williamsia muralis str. 9571414 (99.9%) | ||
| 241 | MH | Rhodococcus sp. str. 5/14 (99.85%) | ||
| 321 | NG | Acinetobacter sp. (100%) | Model species for environmental and biotechnological applications | [40] |
| 343 | MH | Brevibacillus laterosporus str. BL-2 (99.86%) | Invertebrate pathogen | [41] |
| 354 | NG | Variovorax paradoxus str. rif200835 (99.72%) | Plant growth promotion | [42,43,44,45] |
| 362 | NG | Variovorax sp. A2 (100%) | ||
| 364 | NG | Arthrobacter nicotinovorans (100%) | Herbicide degradation (Atrazine) | [46] |
| 365 | NG | Rhodococcus sp. str. 5/14 (99.71%) |
| Sampling Date | Growth Stage of Canola Crop |
|---|---|
| 18 May 2016 | planting |
| 11 July 2016 | pre-flowering |
| 6 October 2016 | flowering |
| 7 November 2016 | post-flowering |
| 8 December 2016 | harvest |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gurusinghe, S.; Brooks, T.L.; Barrow, R.A.; Zhu, X.; Thotagamuwa, A.; Dennis, P.G.; Gupta, V.V.S.R.; Vanniasinkam, T.; Weston, L.A. Technologies for the Selection, Culture and Metabolic Profiling of Unique Rhizosphere Microorganisms for Natural Product Discovery. Molecules 2019, 24, 1955. https://doi.org/10.3390/molecules24101955
Gurusinghe S, Brooks TL, Barrow RA, Zhu X, Thotagamuwa A, Dennis PG, Gupta VVSR, Vanniasinkam T, Weston LA. Technologies for the Selection, Culture and Metabolic Profiling of Unique Rhizosphere Microorganisms for Natural Product Discovery. Molecules. 2019; 24(10):1955. https://doi.org/10.3390/molecules24101955
Chicago/Turabian StyleGurusinghe, Saliya, Tabin L. Brooks, Russell A. Barrow, Xiaocheng Zhu, Agasthya Thotagamuwa, Paul G. Dennis, Vadakattu V. S. R. Gupta, Thiru Vanniasinkam, and Leslie A. Weston. 2019. "Technologies for the Selection, Culture and Metabolic Profiling of Unique Rhizosphere Microorganisms for Natural Product Discovery" Molecules 24, no. 10: 1955. https://doi.org/10.3390/molecules24101955






