High-Frequency Occurrence of Surfactin Monomethyl Isoforms in the Ferment Broth of a Bacillus subtilis Strain Revealed by Ion Trap Mass Spectrometry
Abstract
:1. Introduction
2. Results and Discussions
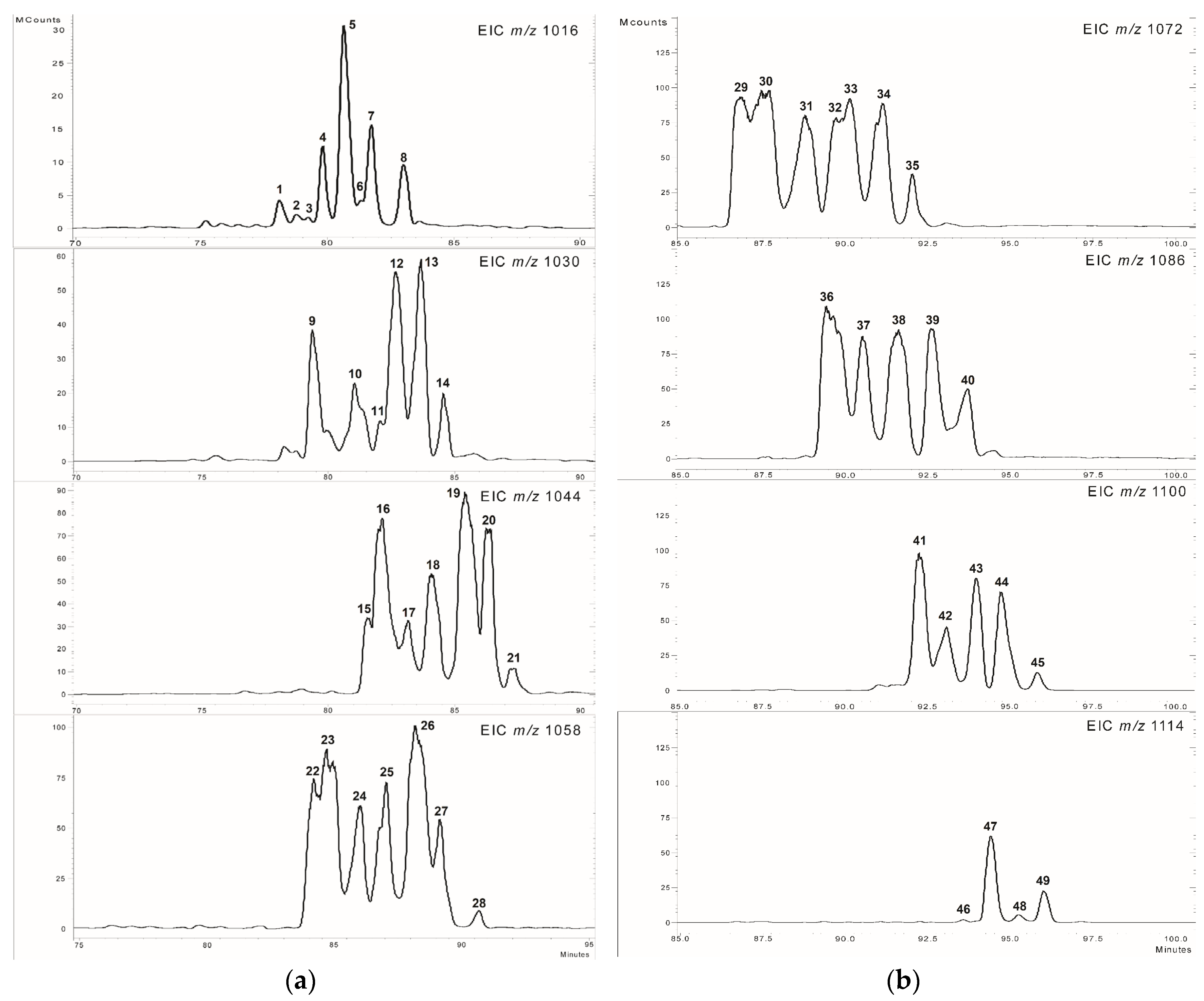
2.1. Separation of Lipopeptides
2.2. Analysis of the MS2 Spectra of Surfactins Over m/z 1058
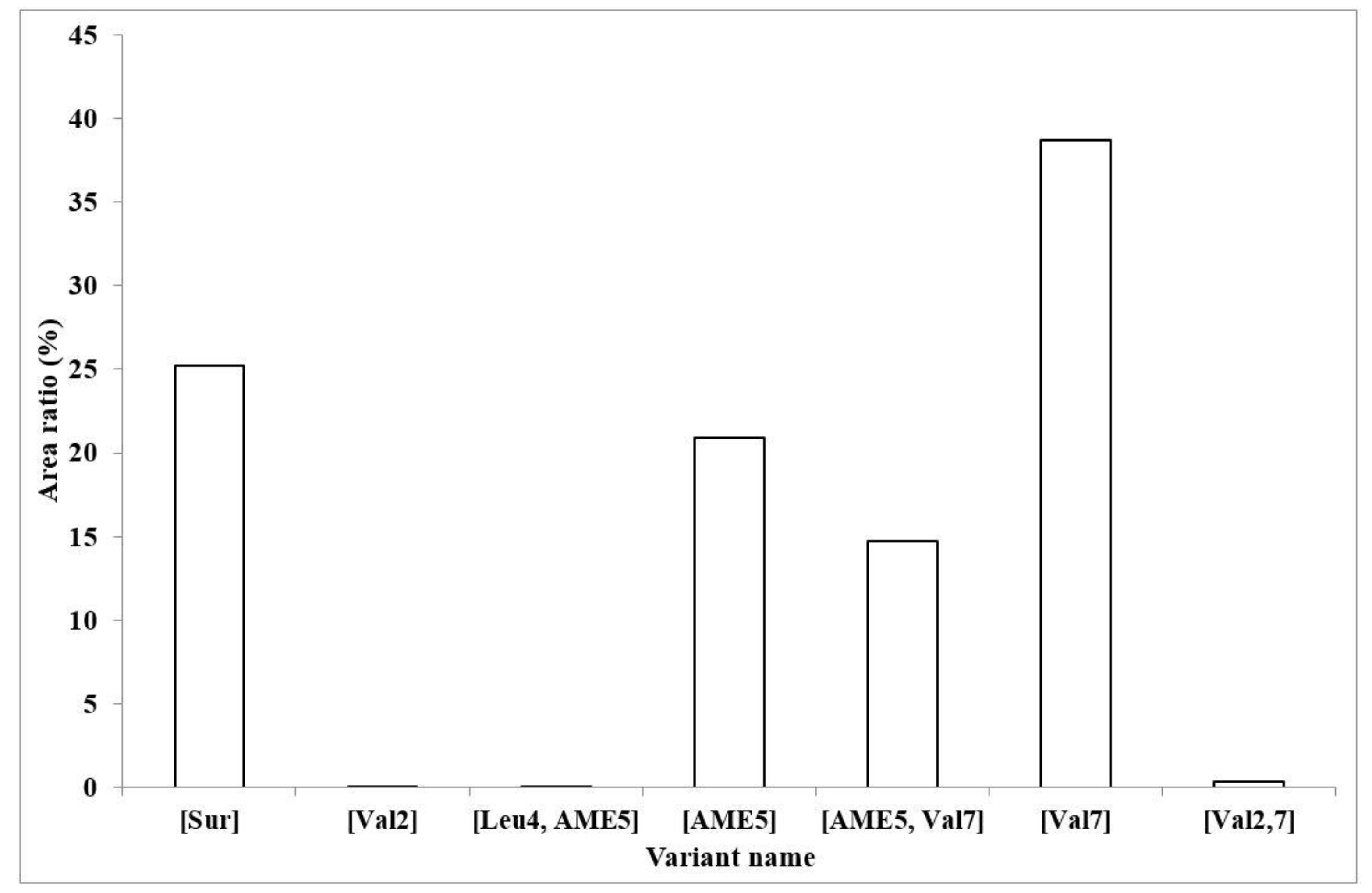
2.3. Characterization of the Amount of Produced Surfactin
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. Microorganism and Culture Conditions
3.3. Extraction of Surfactins
3.4. Liquid Chromatography–Mass Spectrometry Analysis
3.5. Nomenclature
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Stein, T. Bacillus subtilis antibiotics: Structures, syntheses and specific functions. Mol. Microbiol. 2005, 56, 845–857. [Google Scholar] [CrossRef] [PubMed]
- Bonmatin, J.-M.; Laprévote, O.; Peypoux, F. Diversity among microbial cyclic lipopeptides: Iturins and surfactins. Activity-structure relationships to design new bioactive agents. Comb. Chem. High Throughput Screen. 2003, 6, 541–556. [Google Scholar] [CrossRef] [PubMed]
- Pecci, Y.; Rivardo, F.; Martinotti, M.G.; Allegrone, G. LC/ESI-MS/MS characterisation of lipopeptide biosurfactants produced by the Bacillus licheniformis v9t14 strain. J. Mass Spectrom. 2010, 4, 772–778. [Google Scholar] [CrossRef] [PubMed]
- Zou, A.; Liu, J.; Garamus, V.M.; Zheng, K.; Willumeit, R.; Mu, B. Interaction between the natural lipopeptide [Glu1, Asp5] surfactin-C15 and hemoglobin in aqueous solution. Biomacromolecules 2010, 11, 593–599. [Google Scholar] [CrossRef] [PubMed]
- Mulligan, C.N. Environmental applications for biosurfactants. Environ. Pollut. 2005, 133, 183–198. [Google Scholar] [CrossRef] [PubMed]
- Bóka, B.; Manczinger, L.; Kecskeméti, A.; Chandrasekaran, M.; Kadaikunnan, S.; Alharbi, N.S.; Vágvölgyi, C.; Szekeres, A. Ion trap mass spectrometry of surfactins produced by Bacillus subtilis SZMC 6179J reveals novel fagmentation features of cyclic lipopeptides. Rapid Commun. Mass Spectrom. 2016, 30, 1581–1590. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.-S.; Gao, H.; Hong, K.; Yu, J.; Jiang, M.-M.; Lin, H.-P.; Ye, W.-C.; Yao, X.-S. Complete assignments of 1H and 13C NMR spectral data of nine surfactin isomers. Magn. Reson. Chem. 2007, 45, 792–796. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.-S.; Zhao, F.; Gao, H.; Dai, Y.; Yao, Z.-H.; Hong, K.; Li, J.; Ye, W.-C.; Yao, X.-S. Characterization and online detection of surfactin isomers based on HPLC-MSn analyses and their inhibitory effects on the overproduction of nitric oxide and the release of TNF-α and IL-6 in LPS-induced macrophages. Mar. Drugs 2010, 8, 2605–2618. [Google Scholar] [CrossRef] [PubMed]
- Moro, G.V.; Almeida, R.T.R.; Napp, A.P.; Porto, C.; Pilau, E.J.; Lüdtke, D.S.; Moro, A.V.; Vainstein, M.H. Identification and ultra-high-performance liquid chromatography coupled with high-resolution mass spectrometry characterization of biosurfactants, including a new surfactin, isolated from oil-contaminated environments. Microb. Biotechnol. 2018, 11, 759–769. [Google Scholar] [CrossRef] [PubMed]
- Kakinuma, A.; Ouchida, A.; Shima, T.; Sugino, H.; Isono, M.; Tamura, G.; Arima, K. Confirmation of the structure of surfactin by mass spectrometry. Agric. Biol. Chem. 1969, 33, 1669–1671. [Google Scholar] [CrossRef]
- Peypoux, F.; Bonmatin, J.-M.; Labbé, H.; Grangemard, I.; Das, B.C.; Ptak, M.; Wallach, J.; Michel, G. [Ala4] surfactin, a novel isoform from Bacillus subtilis studied by mass and NMR spectroscopies. Eur. J. Biochem. 1994, 224, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Bonmatin, J.-M.; Labbé, H.; Grangemard, I.; Peypoux, F.; Maget-Dana, R.; Ptak, M.; Michel, G. Production, isolation and characterization of [Leu4]- and [Ile4] surfactins from Bacillus subtilis. Lett. Pept. Sci. 1995, 2, 41–47. [Google Scholar] [CrossRef]
- Grangemard, I.; Peypoux, F.; Wallach, J.; Das, B.C.; Labbé, H.; Caille, A.; Genest, M.; Maget-Dana, R.; Ptak, M.; Bonmatin, J.-M. Lipopeptides with improved properties: Structure by NMR, purification by HPLC and structure-activity relationships of new isoleucyl-rich surfactins. J. Pep. Sci. 1997, 3, 145–154. [Google Scholar] [CrossRef]
- Baumgart, F.; Kluge, B.; Ullrich, C.; Vater, J.; Ziessov, D. Identification of amino acid substitutions in the lipopeptide surfactin using 2D NMR spectroscopy. Biochem. Biophys. Res. Commun. 1991, 177, 998–1005. [Google Scholar] [CrossRef]
- Peypoux, F.; Bonmatin, J.-M.; Labbé, H.; Das, B.C.; Ptak, M.; Michel, G. Isolation and characterization of a new variant of surfactin, the [Val7] surfactin. Eur. J. Biochem. 1991, 202, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Kowall, M.; Vater, J.; Kluge, B.; Stein, T.; Franke, P.; Ziessow, D. Separation and characterization of surfactin isoforms produced by Bacillus subtilis OKB 105. J. Colloid Interface Sci. 1998, 204, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Akpa, E.; Jacques, P.; Wathelet, B.; Paquot, M.; Fuchs, R.; Budzikiewicz, H.; Thonart, P. Influence of culture conditions on lipopeptide production by Bacillus subtilis. Appl. Biochem. Biotechnol. 2001, 91–93, 551–561. [Google Scholar] [CrossRef]
- Romano, A.; Vitullo, D.; Di Pietro, A.; Lima, G.; Lanzotti, V. Antifungal lipopeptides from Bacillus amyloliquefaciens strain BO7. J. Nat. Prod. 2011, 74, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.Z.; Liu, X.Y.; Mu, B.Z. The McLafferty rearrangement in the Glu residue in a cyclic lipopeptide determined by QTOF MS/MS. J. Mass Spectrom. 2008, 43, 1673–1678. [Google Scholar] [CrossRef] [PubMed]
- Neta, P.; Pu, Q.L.; Kilpatrick, L.; Yang, X.; Stein, S.E. Dehydration versus deamination of N-terminal glutamine in collision-induced dissociation of protonated peptides. J. Am. Soc. Mass Spectrom. 2007, 18, 27–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourcier, S.; Hoppilliard, Y. Use of diagnostic neutral losses for structural information on unknown aromatic metabolites: An experimental and theoretical study. Rapid Commun. Mass Spectrom. 2009, 23, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Hue, N.; Serani, L.; Laprévote, O. Structural investigation of cyclic peptidolipids from Bacillus subtilis by high-energy tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2001, 15, 203–209. [Google Scholar] [CrossRef]
- Zhao, Y.; Yang, S.Z.; Mu, B.Z. Quantitative analyses of the isoforms of surfactin produced by Bacillus subtilis HSO 121 using GC-MS. Anal. Sci. 2012, 28, 789–793. [Google Scholar] [CrossRef] [PubMed]
- Vágvölgyi, C.; Sajben-Nagy, E.; Bóka, B.; Vörös, M.; Berki, A.; Palágyi, A.; Krisch, J.; Skrbic, B.; Durisic-Mladenovic, N.; Manczinger, L. Isolation and characterization of antagonistic Bacillus strains capable to degrade ethylenethiourea. Curr. Microbiol. 2013, 66, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Besson, F.; Chevanet, C.; Michel, G. Influence of the culture medium on the production of iturin a by Bacillus subtilis. J. Gen. Microbiol. 1987, 133, 767–772. [Google Scholar] [CrossRef] [PubMed]
- Roepstorff, P.; Fohlman, J. Proposal for a common nomenclature for sequence ions in mass spectra of peptides. Biomed. Mass Spectrom. 1984, 11, 601. [Google Scholar] [CrossRef] [PubMed]
- Biemann, K. Sequencing of peptides by tandem mass spectrometry and high-energy collision-induced dissociation. Meth. Enzymol. 1990, 193, 455–479. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |
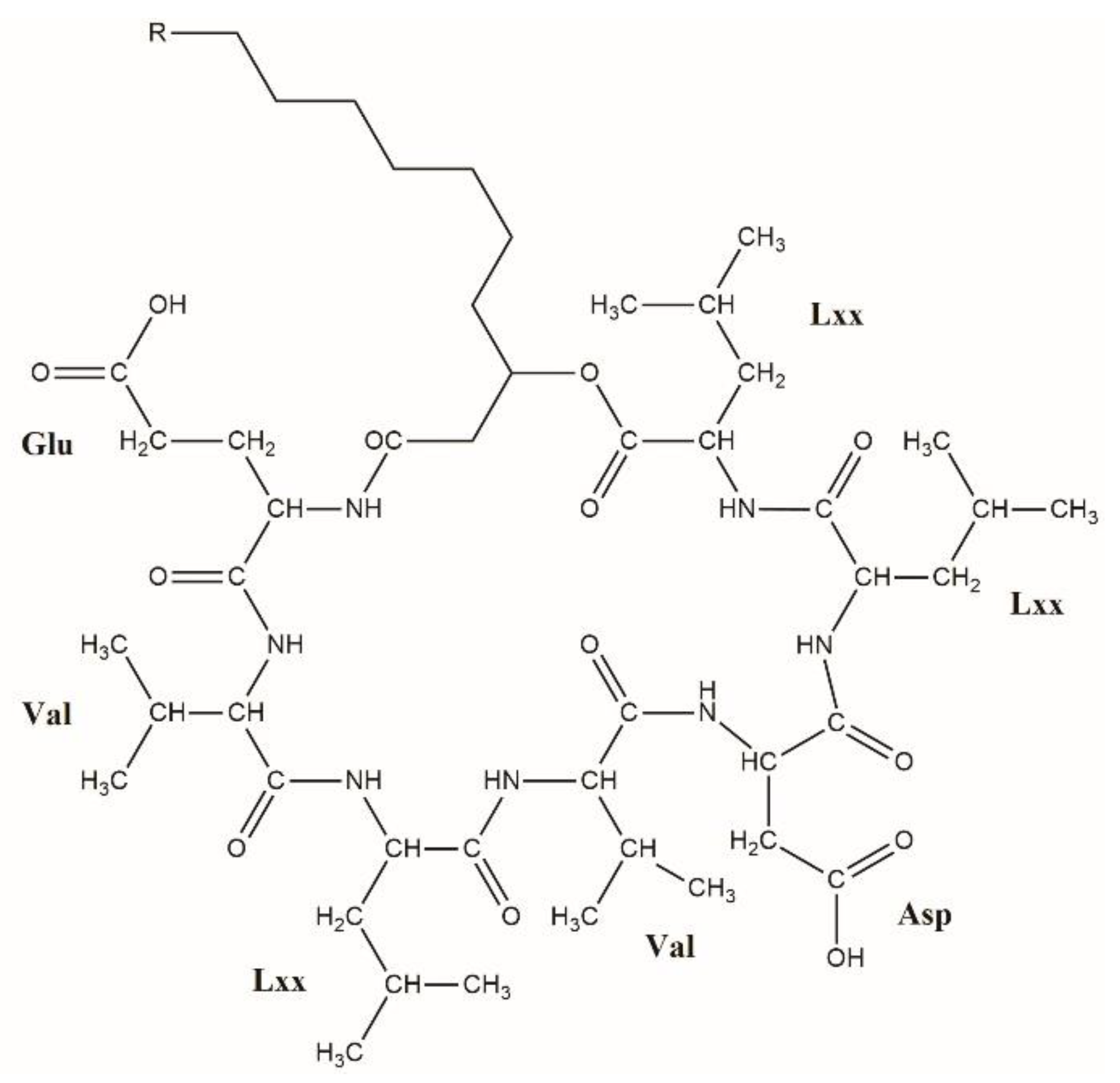




| Nomenclature of Surfactin | Peptide | References | ||||||
|---|---|---|---|---|---|---|---|---|
| AA1 | AA2 | AA3 | AA4 | AA5 | AA6 | AA7 | ||
| [Sur] surfactin | Glu | Leu | Leu | Val | Asp | Leu | Leu | [10] |
| [Val2] surfactin | Glu | Val | Lxx | Val | Asp | Lxx | Lxx | [6] |
| [Ala4] surfactin | Glu | Leu | Leu | Ala | Asp | Leu | Leu | [11] |
| [Leu4] surfactin | Glu | Leu | Leu | Leu | Asp | Leu | Leu | [12] |
| [Ile4] surfactin | Glu | Leu | Leu | Ile | Asp | Leu | Leu | [12] |
| [Ile2,4] surfactin | Glu | Ile | Leu | Ile | Asp | Leu | Leu | [13] |
| [Val7] surfactin | Glu | Leu | Leu | Val | Asp | Leu | Val | [14,15] |
| [Ile7] surfactin | Glu | Leu | Leu | Val | Asp | Leu | Ile | [14] |
| [Val2,7] surfactin | Glu | Val | Leu | Val | Asp | Leu | Val | [13] |
| [Val2, Ile7] surfactin | Glu | Val | Leu | Val | Asp | Leu | Ile | [16] |
| [Ile2, Val7] surfactin | Glu | Ile | Leu | Val | Asp | Leu | Val | [16] |
| [Ile2,4,7] surfactin | Glu | Ile | Leu | Ile | Asp | Leu | Ile | [13] |
| [GME1,AME5] surfactin | GME | Leu | Leu | Val | AME | Leu | Leu | [7] |
| [GME1,Val7] surfactin | GME | Leu | Leu | Val | Asp | Leu | Val | [7] |
| [GME1, Ile7] surfactin | GME | Leu | Leu | Val | Asp | Leu | Ile | [7] |
| [AME5] surfactin | Glu | Leu | Leu | Val | AME | Leu | Leu | [8] |
| [AME5,Val7] surfactin | Glu | Lxx | Lxx | Val | AME | Lxx | Val | This study |
| [Lxx4, AME5] surfactin | Glu | Lxx | Lxx | Lxx | AME | Lxx | Lxx | This study |
| Precursor Ion [M + Na]+ (m/z) | Peak No. | Name | Simple Cleavage | McLafferty Fragments | Internal Fragments | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| b6 | b5 | b50 | b4 | [M+Na]+-72 | b50-72 | y6+H2O | y6b6+H2O | y6b5+H2O | |||
| 1072 | 30 | C16-[Sur] | 959 | 846 | 828 | 731 | 1000 | 756 | 707 | 594 | 481 |
| 34–35 | C17-[Val7] | 973 | 860 | 842 | 745 | 1000 | 770 | 693 | 594 | 481 | |
| 29, 31 | C15-[AME5] | 959 | 846 | 828 | 717 | n.d. | 756 | 721 | 608 | 495 | |
| 32–33 | C16-[AME5,Val7] | 973 | 860 | 842 | 731 | 1000 | n.d. | 707 | 608 | 495 | |
| 1086 | 37, 39 | C17-[Sur] | 973 | 860 | 842 | 745 | 1014 | 770 | 707 | 594 | 481 |
| 36, 38 | C16-[AME5] | 973 | 860 | 842 | 731 | n.d. | n.d. | 721 | 608 | 495 | |
| 40 | C17-[AME5,Val7] | 987 | 874 | 856 | 745 | n.d. | n.d. | 707 | 608 | 495 | |
| 1100 | 41–43 | C17-[AME5] | 987 | 874 | 856 | 745 | n.d. | n.d. | 721 | 608 | 495 |
| 44–45 | C18-[AME5,Val7] | 1001 | 888 | 870 | 759 | n.d. | n.d. | 707 | 608 | 495 | |
| 1114 | 46 | C17-[Lxx4,AME5] | 1001 | 888 | 870 | 759 | n.d. | n.d. | 735 | 622 | 509 |
| 47–49 | C18-[AME5] | 1001 | 888 | 870 | 759 | n.d. | n.d. | 721 | 608 | 495 | |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kecskeméti, A.; Bartal, A.; Bóka, B.; Kredics, L.; Manczinger, L.; Shine, K.; Alharby, N.S.; Khaled, J.M.; Varga, M.; Vágvölgyi, C.; et al. High-Frequency Occurrence of Surfactin Monomethyl Isoforms in the Ferment Broth of a Bacillus subtilis Strain Revealed by Ion Trap Mass Spectrometry. Molecules 2018, 23, 2224. https://doi.org/10.3390/molecules23092224
Kecskeméti A, Bartal A, Bóka B, Kredics L, Manczinger L, Shine K, Alharby NS, Khaled JM, Varga M, Vágvölgyi C, et al. High-Frequency Occurrence of Surfactin Monomethyl Isoforms in the Ferment Broth of a Bacillus subtilis Strain Revealed by Ion Trap Mass Spectrometry. Molecules. 2018; 23(9):2224. https://doi.org/10.3390/molecules23092224
Chicago/Turabian StyleKecskeméti, Anita, Attila Bartal, Bettina Bóka, László Kredics, László Manczinger, Kadaikunnan Shine, Naiyf S. Alharby, Jamal M. Khaled, Mónika Varga, Csaba Vágvölgyi, and et al. 2018. "High-Frequency Occurrence of Surfactin Monomethyl Isoforms in the Ferment Broth of a Bacillus subtilis Strain Revealed by Ion Trap Mass Spectrometry" Molecules 23, no. 9: 2224. https://doi.org/10.3390/molecules23092224





