Comparison of Characteristics and DNAzyme Activity of G4–Hemin Conjugates Obtained via Two Hemin Attachment Methods
Abstract
:1. Introduction
2. Results
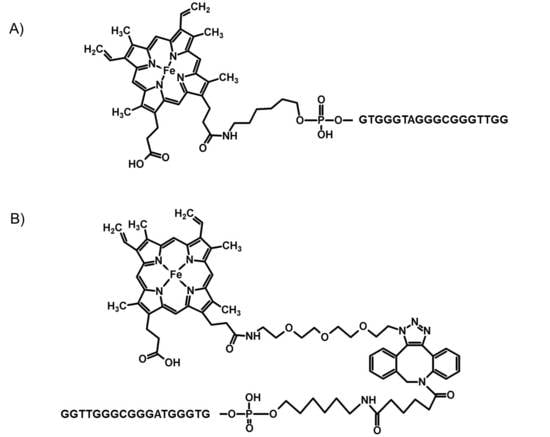
2.1. Chemistry
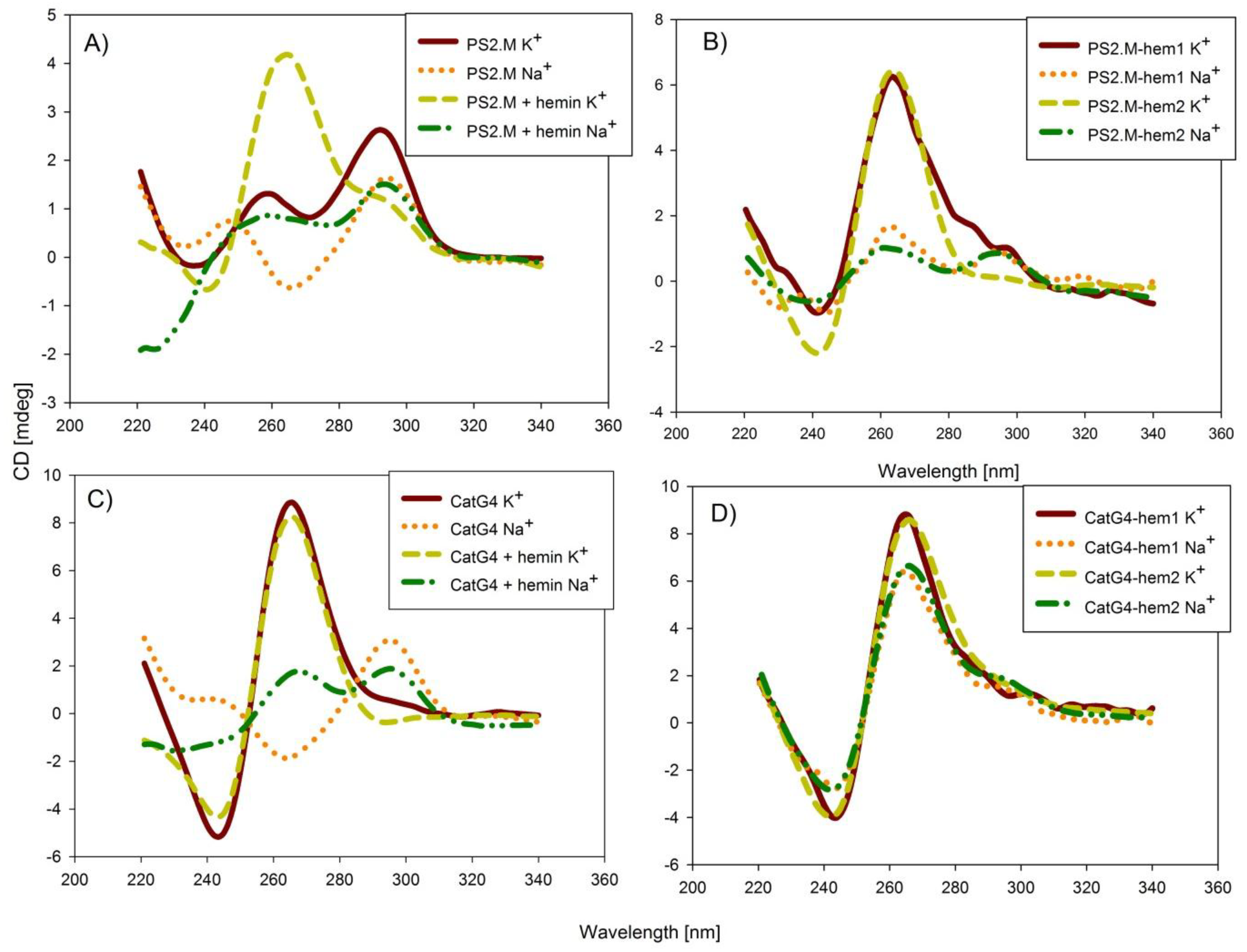
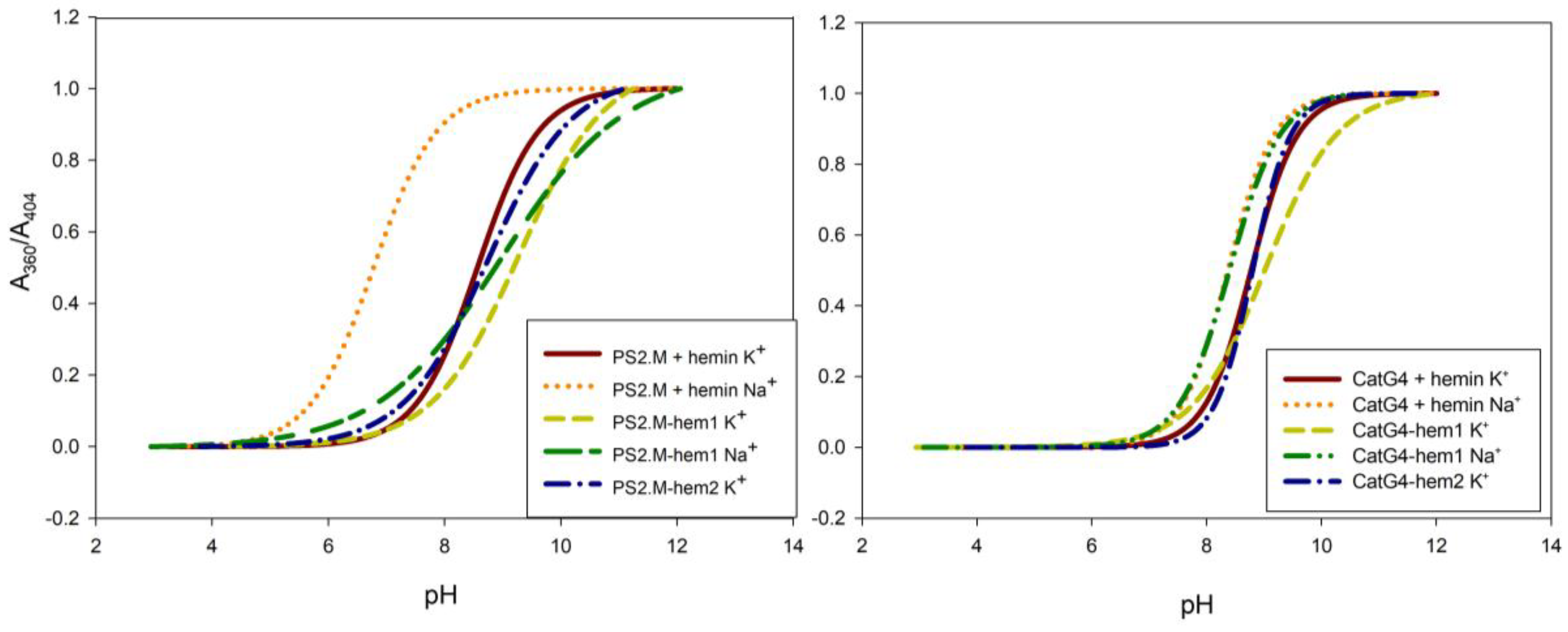
2.2. Spectroscopic Characterization
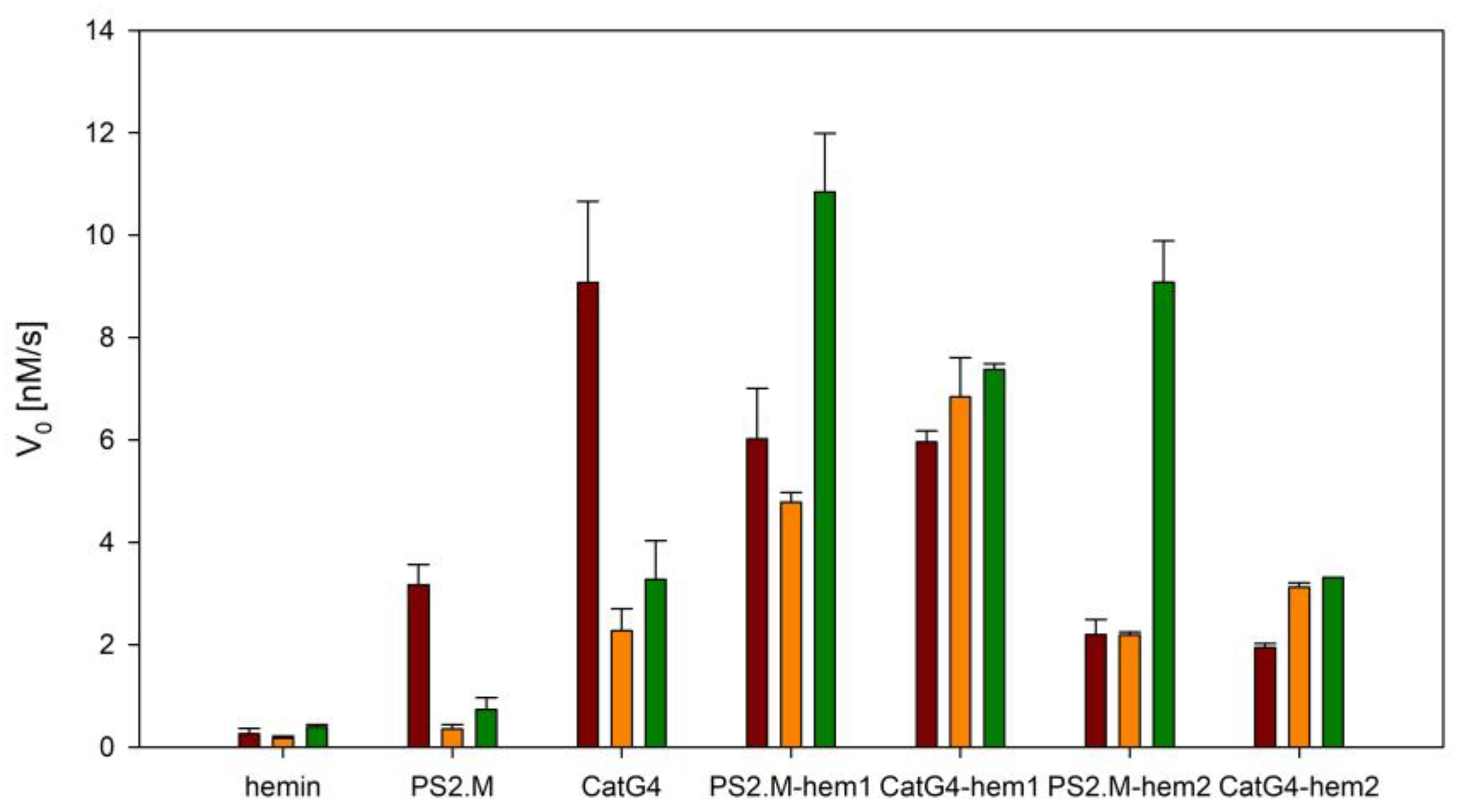
2.3. Activity of DNAzymes
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Circular Dichroism and Thermal Denaturation Experiments
4.3. Spectrophotometric Determination of Hemin pKa
4.4. Peroxidase Activity Measurements
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Travascio, P.; Li, Y.F.; Sen, D. DNA-enhanced peroxidase activity of a DNA aptamer-hemin complex. Chem. Biol. 1998, 5, 505–517. [Google Scholar] [CrossRef]
- Kosman, J.; Juskowiak, B. Peroxidase-mimicking DNAzymes for biosensing applications: A review. Anal. Chim. Acta 2011, 707, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Sen, D.; Poon, L.C.H. RNA and DNA complexes with hemin Fe(III) heme are efficient peroxidases and peroxygenases: How do they do it and what does it mean? Crit. Rev. Biochem. Mol. Biol. 2011, 46, 478–492. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Lu, C.-H.; Willner, I. From cascade catalytic nucleic acids to enzyme-DNA nanostructures: Controlling reactivity, sensing, logic gates operations, and assembly of complex structures. Chem. Rev. 2014, 114, 2881–2941. [Google Scholar] [CrossRef] [PubMed]
- Canale, T.D.; Sen, D. Hemin-utilizing G-quadruplex DNAzymes are strongly active in organic co-solvents. Biochim. Biopbys. Acta Gen. Subj. 2017, 1861, 1455–1462. [Google Scholar] [CrossRef] [PubMed]
- Kong, D.M.; Yang, W.; Wu, J.; Li, C.-X.; Shen, H.-X. Structure-function study of peroxidase-like G-quadruplex-hemin clomplexes. Analyst 2010, 135, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Largy, E.; Mergny, J.L.; Gabelica, V. Role of alkai metal ions in G-quadruplex nucleic acid structure and stability. Met. Ions Life Sci. 2016, 16, 203–258. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, D.; Nakao, A.; Sugimoto, N. Molecular crowding regulates the structural switch of the DNA G-quadruplexes. Biochemistry 2002, 41, 15017–15024. [Google Scholar] [CrossRef] [PubMed]
- Petraccone, L.; Pagano, B.; Giancola, C. Studying the effect of crowding and dehydration on DNA G-quadruplexes. Methods 2012, 57, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Li, Y.; Liu, Z.; Lin, B.; Yi, H.; Xu, F.; Nie, Z.; Yao, S. Insight into G-quadruplex-hemin DNAzyme/RNAzyme: Adjacent adenine as the intermolecular species for remarkable enhancement of enzymatic activity. Nucleic Acids Res. 2016, 44, 7373–7384. [Google Scholar] [CrossRef]
- Chang, T.; Gong, H.; Ding, P.; Liu, X.; Li, W.; Bing, T.; Cao, Z.; Shangguan, D. Activity enhancement of G-quadruplex/hemin DNAzyme by flanking d(CCC). Chemistry 2016, 22, 4015–4021. [Google Scholar] [CrossRef] [PubMed]
- Kosman, J.; Juskowiak, B. Hemin/G-quadruplex structure and activity alteration induced by magnesium cations. Int. J. Biol. Macromol. 2016, 85, 555–564. [Google Scholar] [CrossRef] [PubMed]
- Qui, C.; Zhang, N.; Yan, J.; Liu, X.; Bing, T.; Mei, H.; Shangguan, D. Activity enhancement of G-quadruplex/hemin DNAzyme by spermine. RSC Adv. 2014, 4, 1441–1448. [Google Scholar] [CrossRef]
- Cheng, M.; Zhuo, J.; Jia, G.; Ai, X.; Mergny, J.-L.; Li, C. Relations between the loop transposition of DNA G-quadruplex and the catalytic function of DNAzyme. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 1913–1920. [Google Scholar] [CrossRef] [PubMed]
- Golub, E.; Albada, H.B.; Liao, W.-C.; Biniuri, Y.; Willner, I. Nucleoapzymes: Hemin/G-quadruplex DNAzyme-aptamer binding site conjugates with superior enzyme-like catalytic functions. J. Am. Chem. Soc. 2016, 138, 164–172. [Google Scholar] [CrossRef] [PubMed]
- Gribas, A.V.; Zhao, S.; Sakharov, I.Y. Improved method for chemiluminescent determination of peroxidase-mimicking DNAzyme activity. Anal. Biochem. 2014, 466, 19–23. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhu, K.; Zhu, Z.; Fu, H.; Jenkins, G.; Wang, C.; Zou, Y.; Lu, X.; Yang, C.J. Backbone modification promotes peroxidase activity of G-quadruplex-based DNAzyme. Chem. Commun. 2012, 48, 8347–8349. [Google Scholar] [CrossRef] [PubMed]
- Stefan, L.; Denat, F.; Monchaud, D. Insights into how nucleotide supplements enhance the peroxidase mimicking DNAzyme activity of the G-quadruplex/hemin system. Nucleic Acids Res. 2012, 40, 8759–8772. [Google Scholar] [CrossRef] [PubMed]
- Thirstrup, D.; Baird, G.S. Histochemical application of a peroxidase DNAzyme with a covalently attached hemin cofactor. Anal. Chem. 2010, 82, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, S.; Wang, J.; Sintim, H.O. DNA-Based Peroxidation Catalyst—What Is the Exact Role of Topology on Catalysis and Is There a Special Binding Site for Catalysis? Chem. Eur. J. 2011, 5691–5698. [Google Scholar] [CrossRef] [PubMed]
- Gribas, A.V.; Korolev, S.P.; Zatsepin, T.S.; Gottikh, M.B.; Sakharov, I.Y. Structure–activity relationship study for design of highly active covalent peroxidase-mimicking DNAzyme. RSC Adv. 2015, 5, 51672–51677. [Google Scholar] [CrossRef]
- Kosman, J.; Stanislawska, A.; Gluszynska, A.; Juskowiak, B. Conjugation of hemin to G-quadruplex forming oligonucleotide usingclick chemistry. Int. J. Biol. Macromol. 2017, 101, 799–804. [Google Scholar] [CrossRef] [PubMed]
- Fasman, G.D. (Ed.) Nucleic Acids. In Handbook of Biochemistry and Molecular Biology, 3rd ed.; CRC Press: Boca Raton, FL, USA, 1975; Volume 1, ISBN 9780878195039. [Google Scholar]
- Travascio, P.; Witting, P.K.; Mauk, A.G.; Sen, D. The peroxidase activity of a hemin-DNA oligonucleotide complex: Free radical damage to specific guanines bases of DNA. J. Am. Chem. Soc. 2001, 123, 1337–1348. [Google Scholar] [CrossRef] [PubMed]
- Kachab, E.H.; Wu, W.-Y.; Chapman, C.B. The development of enzyme-linked immunosorbent assay (ELISA) for cephalexin. J. Immunol. Methods 1992, 147, 33–41. [Google Scholar] [CrossRef]
- Yang, X.; Ma, K. Determination of hydrogen peroxide generated by reduced nicotinamide adenine dinucleotide oxidase. Anal. Biochem. 2005, 344, 130–134. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, S.; Sintim, H.O. Investigating the interaction between cations, peroxidation substrates and G-quadruplex topology in DNAzyme peroxidation reaction using statistical testing. Anal. Chim. Acta 2012, 747, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Chen, J.; Cheng, M.; Monchaud, D.; Zhuo, J.; Ju, H. A thermophilic tetramolecular G-quadruplex/hemin DNAzyme. Angew. Chem. Int. Ed. 2017, 56, 16636–16640. [Google Scholar] [CrossRef] [PubMed]
- Kypr, J.; Kejnovska, I.; Renciuk, D.; Vorlickova, M. Circular dichroism nad conformational polymorphism of DNA. Nucleic Acids Res. 2009, 37, 1713–1725. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Not Available. |



| Name | Sequence |
|---|---|
| PS2.M | 5′–GTG GGT AGG GCG GGT TGG–3′ |
| CatG4 | 5′–TGG GTA GGG CGG GTT GGG AAA–3′ |
| PS2.M–hem1 | Hemin–(C6H12)–5′–GTG GGT AGG GCG GGT TGG–3′ |
| PS2.M–hem2 | Hemin–(C35N5O5H49)–5′–GTG GGT AGG GCG GGT TGG–3′ |
| CatG4–hem1 | Hemin–(C6H12)–5′–TGG GTA GGG CGG GTT GGG AAA–3′ |
| CatG4–hem2 | Hemin–(C35N5O5H49)–5′–TGG GTA GGG CGG GTT GGG AAA–3′ |
| Sequence | Tm [°C], 100 mM KCl | Tm [°C], 100 mM NaCl |
|---|---|---|
| PS2.M | 56.5 ± 0.9 | 46.0 ± 0.4 |
| PS2.M/hemin | 64.6 ± 1.0 | 45.2 ± 0.6 |
| PS2.M–hem1 | >80.0 a | >80.0 a |
| PS2.M–hem2 | >80.0 a | 58.4 ± 1.0 |
| CatG4 | 79.2 ± 1.5 | 41.8 ± 0.5 |
| CatG4/hemin | >80.0 a | 42.2 ± 1.4 |
| CatG4–hem2 | >80.0 a | 53.0 ± 1.9 |
| CatG4–hem1 | >80.0 a | >80.0 a |
| >80.0 a,b | >80.0 a,b | |
| >80.0 a,c | 79.6 ± 0.3 c | |
| 85.2 ± 1.2 d | 73.0 ± 0.4 d | |
| 64.0 ± 0.7 e | 64.0 ± 0.7 e |
| Sequence | K+ | Na+ |
|---|---|---|
| PS2.M/hemin | >9 1 | 6.9 ± 0.2 |
| PS2.M–hem1 | 9.2 ± 0.1 | 9.1 ± 0.2 |
| PS2.M–hem2 | 8.7 ± 0.1 | Nd |
| CatG4/hemin | >9 1 | 8.5 ± 0.2 |
| CatG4–hem1 | 8.8 ± 0.2 | 8.6 ± 0.3 |
| CatG4–hem2 | 8.7 ± 0.3 | Nd |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kosman, J.; Żukowski, K.; Juskowiak, B. Comparison of Characteristics and DNAzyme Activity of G4–Hemin Conjugates Obtained via Two Hemin Attachment Methods. Molecules 2018, 23, 1400. https://doi.org/10.3390/molecules23061400
Kosman J, Żukowski K, Juskowiak B. Comparison of Characteristics and DNAzyme Activity of G4–Hemin Conjugates Obtained via Two Hemin Attachment Methods. Molecules. 2018; 23(6):1400. https://doi.org/10.3390/molecules23061400
Chicago/Turabian StyleKosman, Joanna, Krzysztof Żukowski, and Bernard Juskowiak. 2018. "Comparison of Characteristics and DNAzyme Activity of G4–Hemin Conjugates Obtained via Two Hemin Attachment Methods" Molecules 23, no. 6: 1400. https://doi.org/10.3390/molecules23061400