Non-Destructive and Rapid Variety Discrimination and Visualization of Single Grape Seed Using Near-Infrared Hyperspectral Imaging Technique and Multivariate Analysis
Abstract
:1. Introduction
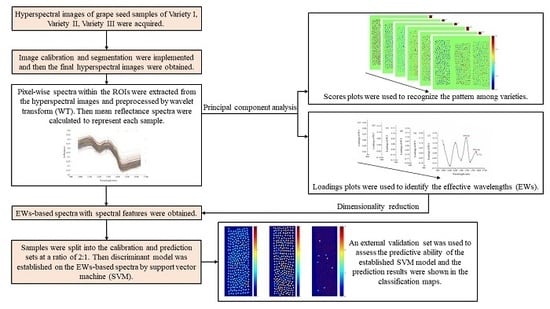
2. Results
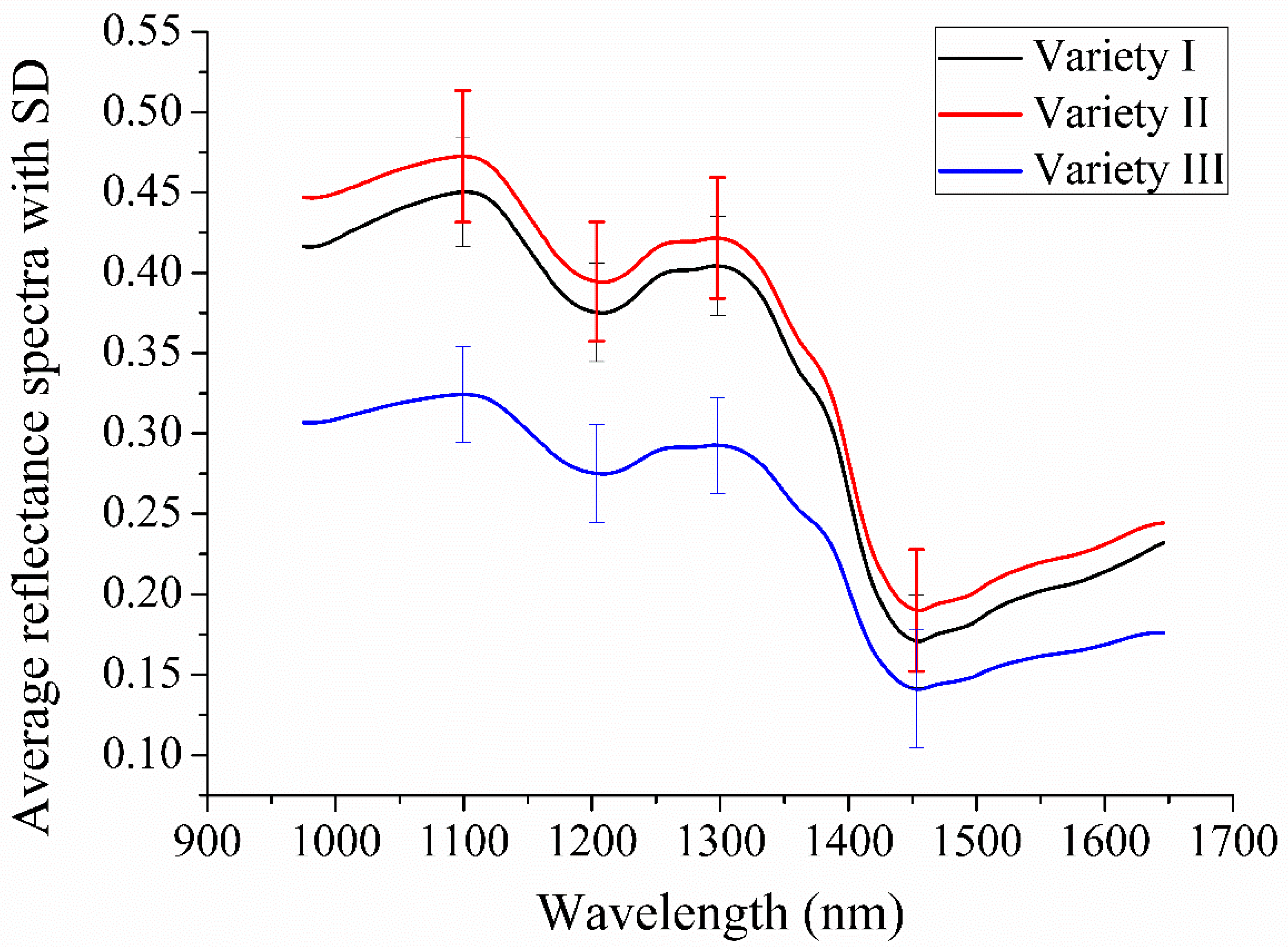
2.1. Overview of Spectral Profiles
2.2. Principal Component Analysis
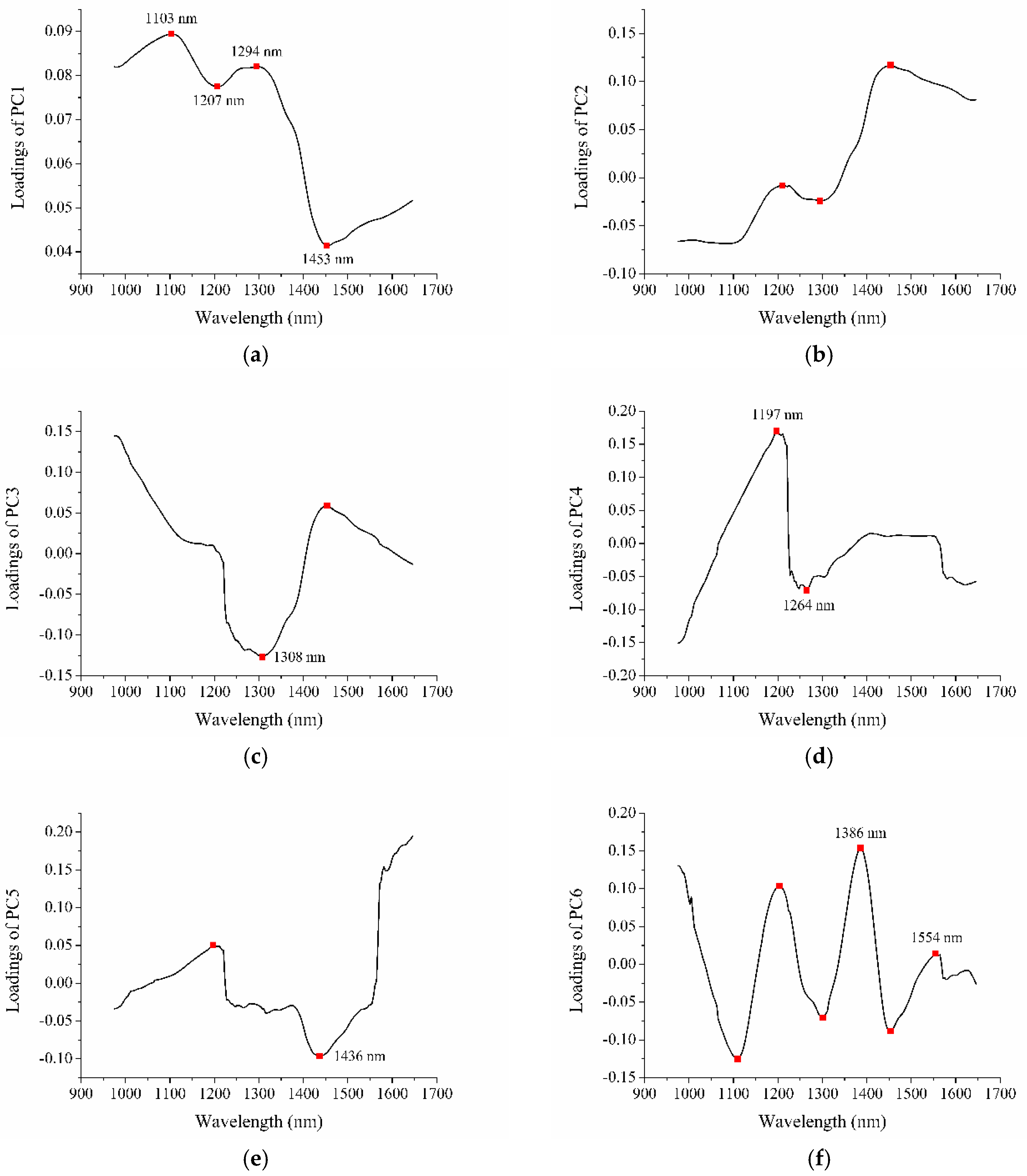
2.3. Effective Wavelength Selection
2.4. Discriminant Models Based on the Effective Wavelengths
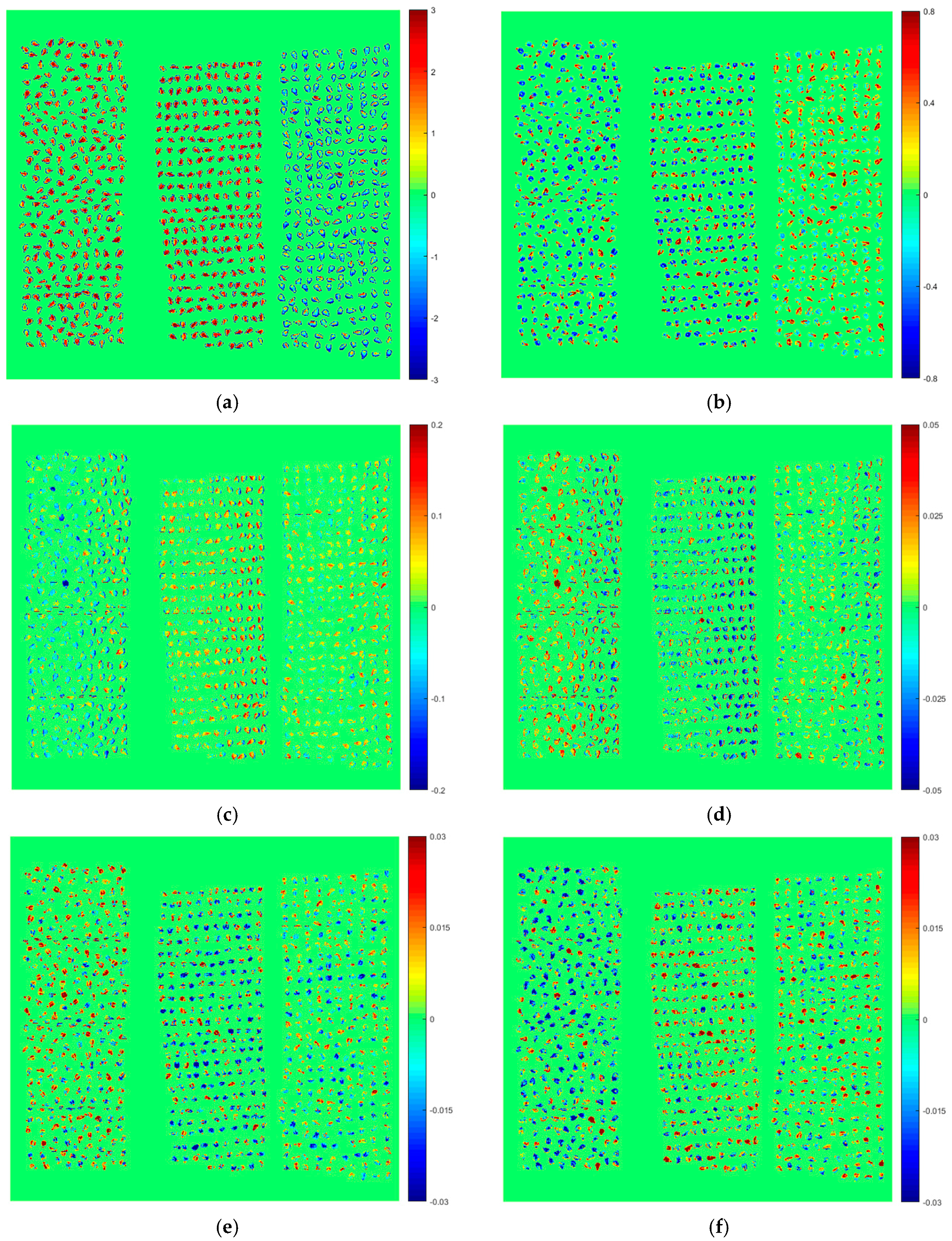
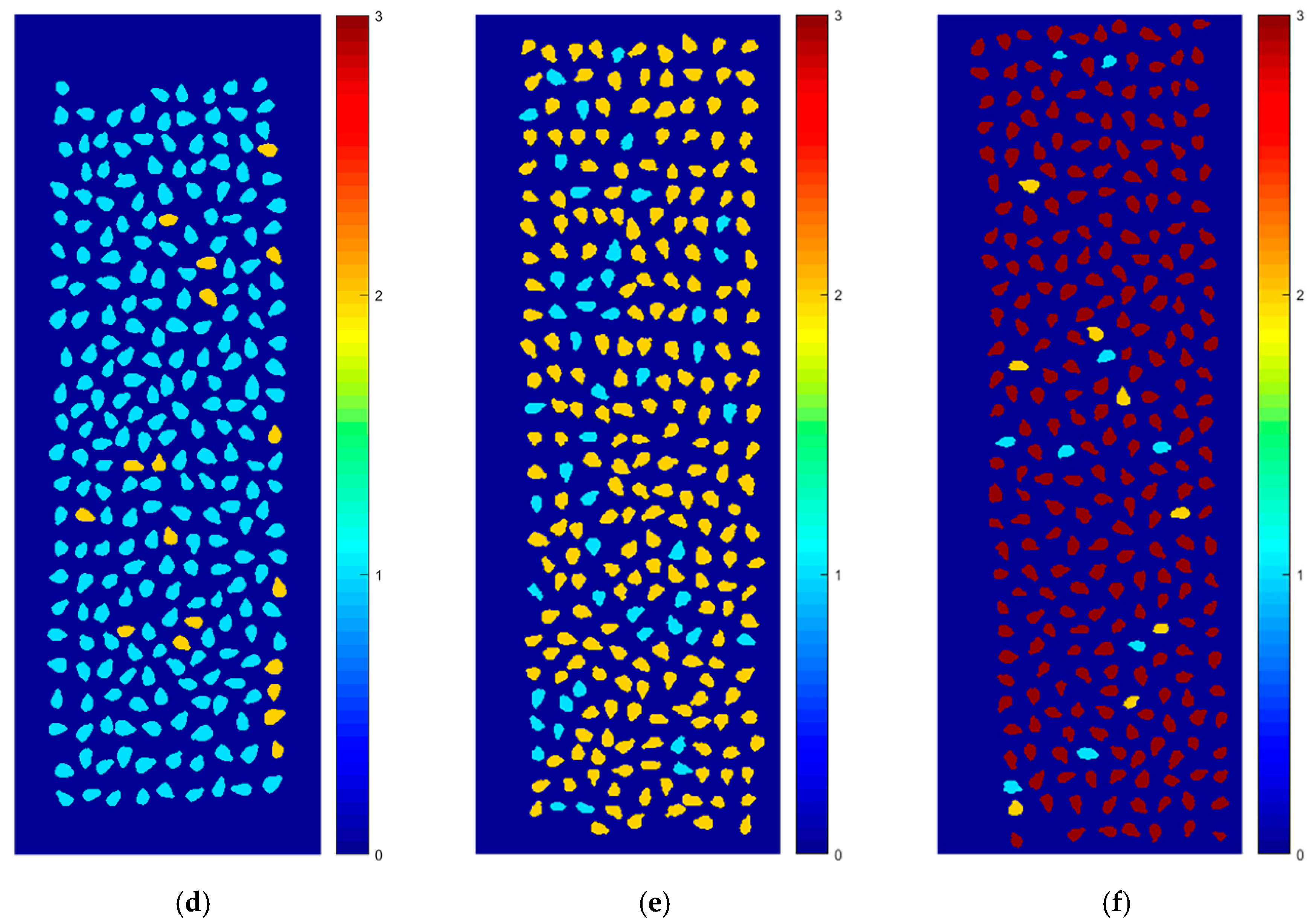
2.5. Varieties’ Classification and Visualization of External Validation Images
3. Discussion
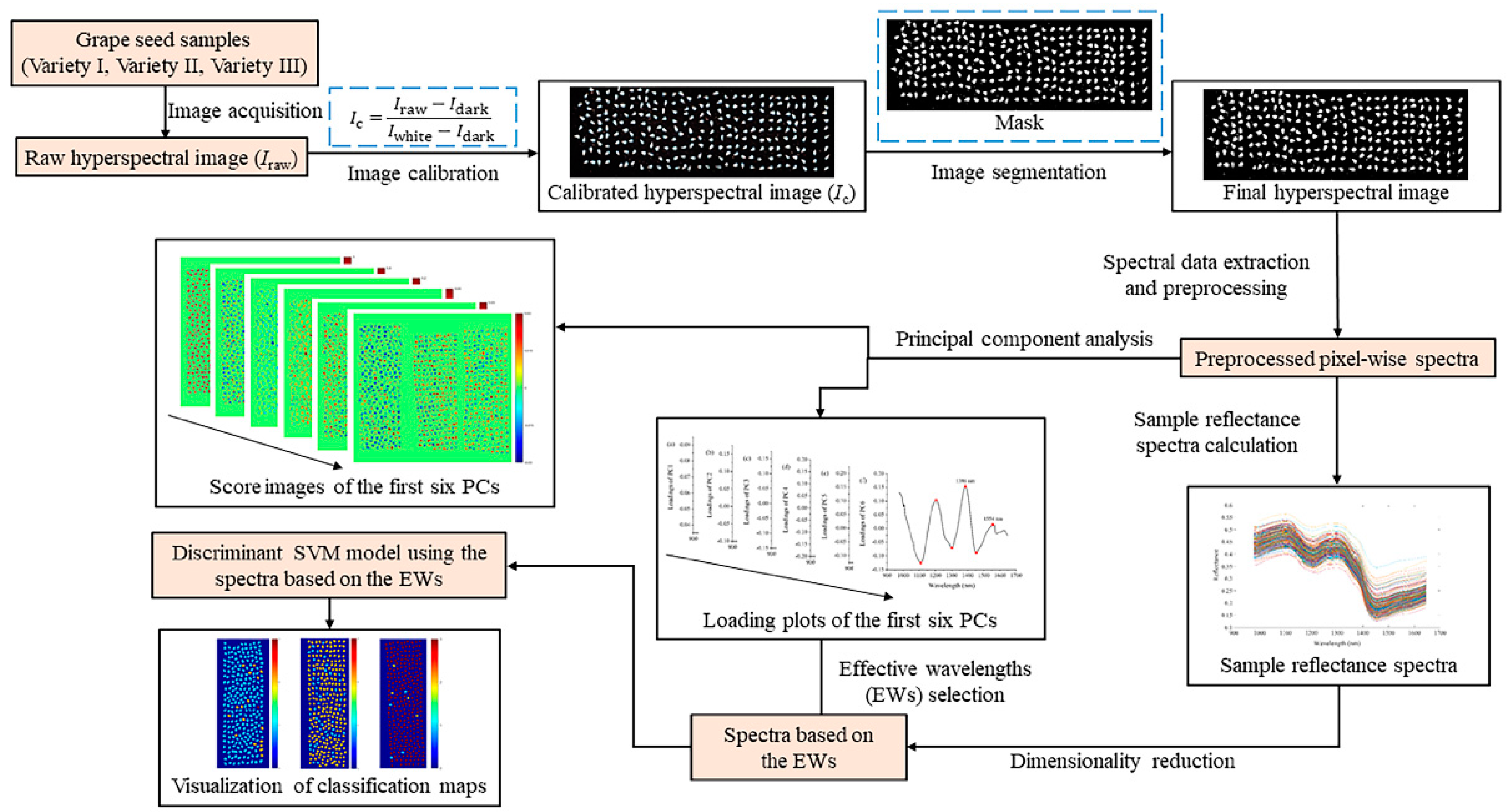
4. Materials and Methods
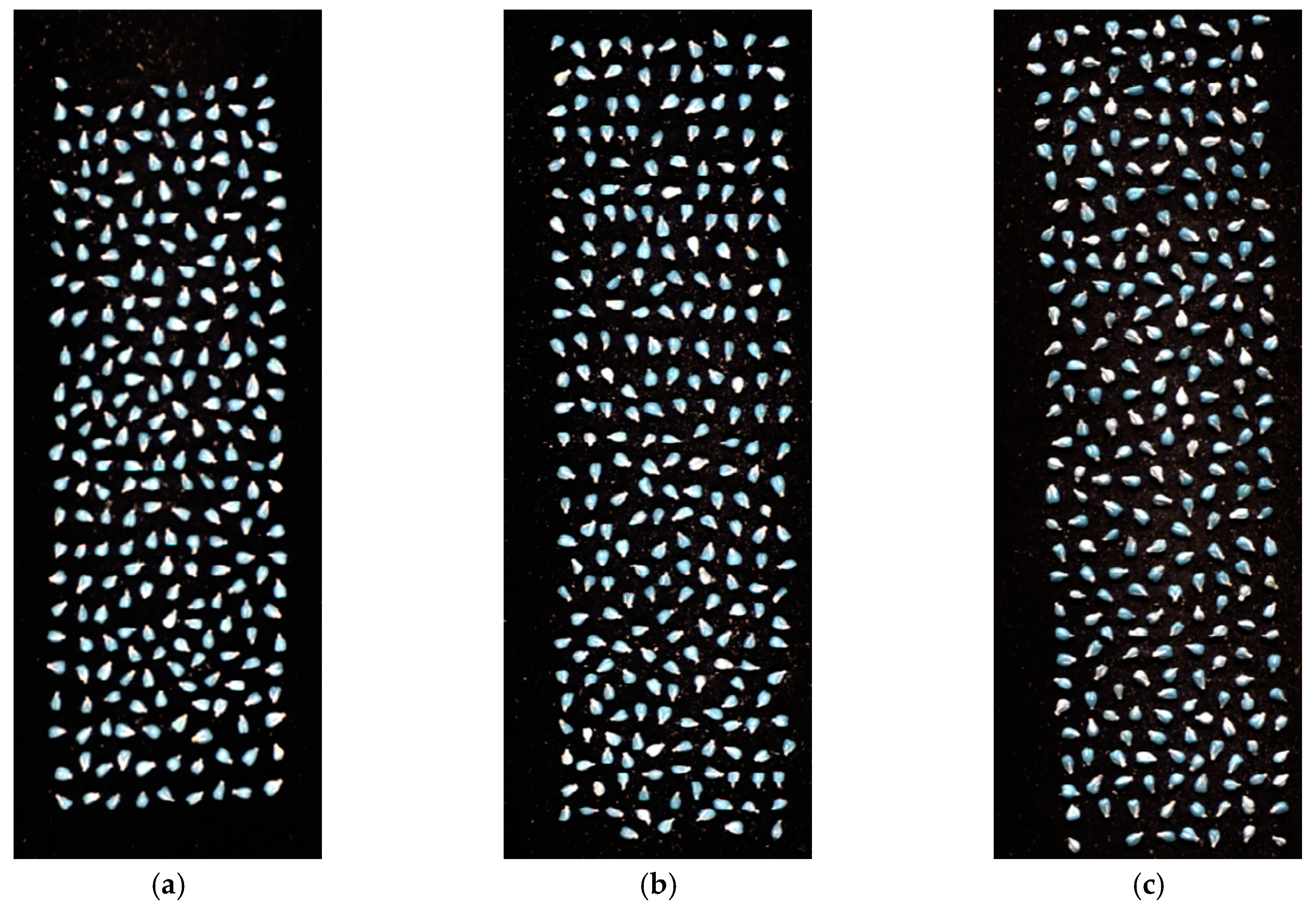
4.1. Sample Preparation
4.2. Hyperspectral Image Acquisition
4.2.1. Hyperspectral Imaging System
4.2.2. Hyperspectral Imaging Acquisition and Correction
4.3. Data Analysis
4.3.1. Data Extraction and Preprocessing
4.3.2. Sample Set Split
4.3.3. Principal Component Analysis
4.3.4. Discriminant Method
4.3.5. Software
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- OIV (2014). Available online: http://www.oiv.int/en/databases-and-statistics/statistics (accessed on 20 April 2018).
- Casazza, A.A.; Aliakbarian, B.; Mantegna, S.; Cravotto, G.; Perego, P. Extraction of phenolics from Vitis vinifera wastes using non-conventional techniques. J. Food Eng. 2010, 100, 50–55. [Google Scholar] [CrossRef]
- Peixoto, C.M.; Dias, M.I.; Alves, M.J.; Calhelha, R.C.; Barros, L.; Pinho, S.P.; Ferreira, I. Grape pomace as a source of phenolic compounds and diverse bioactive properties. Food Chem. 2018, 253, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Nogales-Bueno, J.; Baca-Bocanegra, B.; Rooney, A.; Hernandez-Hierro, J.M.; Byrne, H.J.; Heredia, F.J. Study of phenolic extractability in grape seeds by means of ATR-FTIR and Raman spectroscopy. Food Chem. 2017, 232, 602–609. [Google Scholar] [CrossRef] [PubMed]
- Cadot, Y.; Minana-Castello, M.T.; Chevalier, M. Anatomical, histological, and histochemical changes in grape seeds from Vitis vinifera L. cv cabernet franc during fruit development. J. Agric. Food Chem. 2006, 54, 9206–9215. [Google Scholar] [CrossRef] [PubMed]
- Obreque-Slier, E.; Pena-Neira, A.; Lopez-Solis, R.; Zamora-Marin, F.; Ricardo-da Silva, J.M.; Laureano, O. Comparative study of the phenolic composition of seeds and skins from Carmenere and Cabernet Sauvignon grape varieties (Vitis vinifera L.) during ripening. J. Agric. Food Chem. 2010, 58, 3591–3599. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez Montealegre, R.; Romero Peces, R.; Chacón Vozmediano, J.L.; Martínez Gascueña, J.; García Romero, E. Phenolic compounds in skins and seeds of ten grape Vitis vinifera varieties grown in a warm climate. J. Food Compos. Anal. 2006, 19, 687–693. [Google Scholar] [CrossRef]
- Xu, C.; Zhang, Y.; Cao, L.; Lu, J. Phenolic compounds and antioxidant properties of different grape cultivars grown in China. Food Chem. 2010, 119, 1557–1565. [Google Scholar] [CrossRef]
- Lachman, J.; Hejtmánková, A.; Hejtmánková, K.; Horníčková, Š.; Pivec, V.; Skala, O.; Dědina, M.; Přibyl, J. Towards complex utilisation of winemaking residues: Characterisation of grape seeds by total phenols, tocols and essential elements content as a by-product of winemaking. Ind. Crop. Prod. 2013, 49, 445–453. [Google Scholar] [CrossRef]
- Orrù, M.; Grillo, O.; Lovicu, G.; Venora, G.; Bacchetta, G. Morphological characterisation of Vitis vinifera L. seeds by image analysis and comparison with archaeological remains. Veg. Hist. Archaeobot. 2013, 22, 231–242. [Google Scholar] [CrossRef]
- Mao, R.; Xia, P.; He, Z.; Liu, Y.; Liu, F.; Zhao, H.; Han, R.; Liang, Z. Identification of seeds based on molecular markers and secondary metabolites in Senna obtusifolia and Senna occidentalis. Bot. Stud. 2017, 58, 43. [Google Scholar] [CrossRef] [PubMed]
- ElMasry, G.; Sun, D.-W.; Allen, P. Near-infrared hyperspectral imaging for predicting colour, pH and tenderness of fresh beef. J. Food Eng. 2012, 110, 127–140. [Google Scholar] [CrossRef]
- Zhang, C.; Guo, C.; Liu, F.; Kong, W.; He, Y.; Lou, B. Hyperspectral imaging analysis for ripeness evaluation of strawberry with support vector machine. J. Food Eng. 2016, 179, 11–18. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhu, S.; Zhang, C.; Feng, X.; Feng, L.; He, Y. Application of hyperspectral imaging and chemometrics for variety classification of maize seeds. RSC Adv. 2018, 8, 1337–1345. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Li, X.; Zhu, F.; He, Y. Application of hyperspectral imaging technology to discriminate different geographical origins of Jatropha curcas L. seeds. Comput. Electron. Agric. 2013, 99, 186–193. [Google Scholar] [CrossRef]
- Kandpal, L.M.; Lohumi, S.; Kim, M.S.; Kang, J.; Cho, B. Near-infrared hyperspectral imaging system coupled with multivariate methods to predict viability and vigor in muskmelon seeds. Sensor. Actuat. B-Chem. 2016, 229, 534–544. [Google Scholar] [CrossRef]
- Lee, H.; Kim, M.S.; Song, Y.R.; Oh, C.S.; Lim, H.S.; Lee, W.H.; Kang, J.S.; Cho, B.K. Non-destructive evaluation of bacteria-infected watermelon seeds using visible/near-infrared hyperspectral imaging. J. Sci. Food Agric. 2017, 97, 1084–1092. [Google Scholar] [CrossRef] [PubMed]
- Williams, P.J.; Geladi, P.; Britz, T.J.; Manley, M. Investigation of fungal development in maize kernels using NIR hyperspectral imaging and multivariate data analysis. J. Cereal Sci. 2012, 55, 272–278. [Google Scholar] [CrossRef]
- Cen, H.; He, Y. Theory and application of near infrared reflectance spectroscopy in determination of food quality. Trends Food Sci. Technol. 2007, 18, 72–83. [Google Scholar] [CrossRef]
- Khodabux, K.; L’Omelette, M.S.; Jhaumeer-Laulloo, S.; Ramasami, P.; Rondeau, P. Chemical and near-infrared determination of moisture, fat and protein in tuna fishes. Food Chem. 2007, 102, 669–675. [Google Scholar] [CrossRef]
- Su, W.; Sun, D. Potential of hyperspectral imaging for visual authentication of sliced organic potatoes from potato and sweet potato tubers and rapid grading of the tubers according to moisture proportion. Comput. Electron. Agric. 2016, 125, 113–124. [Google Scholar] [CrossRef]
- Ribeiro, J.S.; Ferreira, M.M.; Salva, T.J. Chemometric models for the quantitative descriptive sensory analysis of Arabica coffee beverages using near infrared spectroscopy. Talanta 2011, 83, 1352–1358. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Pulido, F.J.; Barbin, D.F.; Sun, D.; Gordillo, B.; González-Miret, M.L.; Heredia, F.J. Grape seed characterization by NIR hyperspectral imaging. Postharvest Biol. Technol. 2013, 76, 74–82. [Google Scholar] [CrossRef]
- Kong, W.; Zhang, C.; Huang, W.; Liu, F.; He, Y. Application of hyperspectral imaging to detect Sclerotinia sclerotiorum on oilseed rape stems. Sensors 2018, 18, 123. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Jiang, H.; Liu, F.; He, Y. Application of near-infrared hyperspectral imaging with variable selection methods to determine and visualize caffeine content of coffee beans. Food Bioprocess Technol. 2017, 10, 213–221. [Google Scholar] [CrossRef]
- Rodríguez-Pulido, F.J.; Hernández-Hierro, J.M.; Nogales-Bueno, J.; Gordillo, B.; González-Miret, M.L.; Heredia, F.J. A novel method for evaluating flavanols in grape seeds by near infrared hyperspectral imaging. Talanta 2014, 122, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Chu, B.; He, Y. Prediction of banana color and firmness using a novel wavelengths selection method of hyperspectral imaging. Food Chem. 2018, 245, 132–140. [Google Scholar] [CrossRef] [PubMed]
- Serranti, S.; Gargiulo, A.; Bonifazi, G. Hyperspectral imaging for process and quality control in recycling plants of polyolefin flakes. J. Near Infrared Spectrosc. 2012, 20, 573–581. [Google Scholar] [CrossRef]
- Dong, J.J.; Li, Q.L.; Yin, H.; Zhong, C.; Hao, J.G.; Yang, P.F.; Tian, Y.H.; Jia, S.R. Predictive analysis of beer quality by correlating sensory evaluation with higher alcohol and ester production using multivariate statistics methods. Food Chem. 2014, 161, 376–382. [Google Scholar] [CrossRef] [PubMed]
- Devos, O.; Downey, G.; Duponchel, L. Simultaneous data pre-processing and SVM classification model selection based on a parallel genetic algorithm applied to spectroscopic data of olive oils. Food Chem. 2014, 148, 124–130. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |
| Calibration Set (28,290 Samples) | Prediction Set (14,267 Samples) | |||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | Accuracy | 1 | 2 | Accuracy | ||
| 1 | 8355 | 742 | 23 | 91.6% | 4020 | 592 | 28 | 86.6% |
| 2 | 760 | 8495 | 54 | 91.3% | 947 | 3767 | 10 | 79.7% |
| 3 | 11 | 23 | 9827 | 99.7% | 27 | 13 | 4863 | 99.2% |
| Total | 94.3% | 88.7% | ||||||
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Y.; Zhang, C.; Zhu, S.; Gao, P.; Feng, L.; He, Y. Non-Destructive and Rapid Variety Discrimination and Visualization of Single Grape Seed Using Near-Infrared Hyperspectral Imaging Technique and Multivariate Analysis. Molecules 2018, 23, 1352. https://doi.org/10.3390/molecules23061352
Zhao Y, Zhang C, Zhu S, Gao P, Feng L, He Y. Non-Destructive and Rapid Variety Discrimination and Visualization of Single Grape Seed Using Near-Infrared Hyperspectral Imaging Technique and Multivariate Analysis. Molecules. 2018; 23(6):1352. https://doi.org/10.3390/molecules23061352
Chicago/Turabian StyleZhao, Yiying, Chu Zhang, Susu Zhu, Pan Gao, Lei Feng, and Yong He. 2018. "Non-Destructive and Rapid Variety Discrimination and Visualization of Single Grape Seed Using Near-Infrared Hyperspectral Imaging Technique and Multivariate Analysis" Molecules 23, no. 6: 1352. https://doi.org/10.3390/molecules23061352