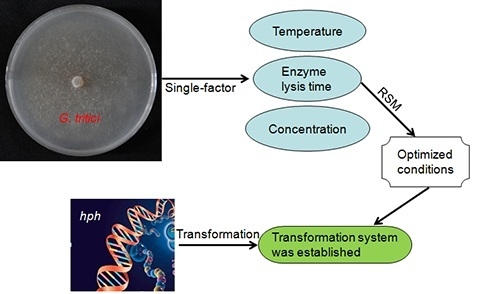
Optimization of Production Conditions for Protoplasts and Polyethylene Glycol-Mediated Transformation of Gaeumannomyces tritici
Abstract
:1. Introduction
2. Results
2.1. Effects of Enzyme on Protoplast Yield
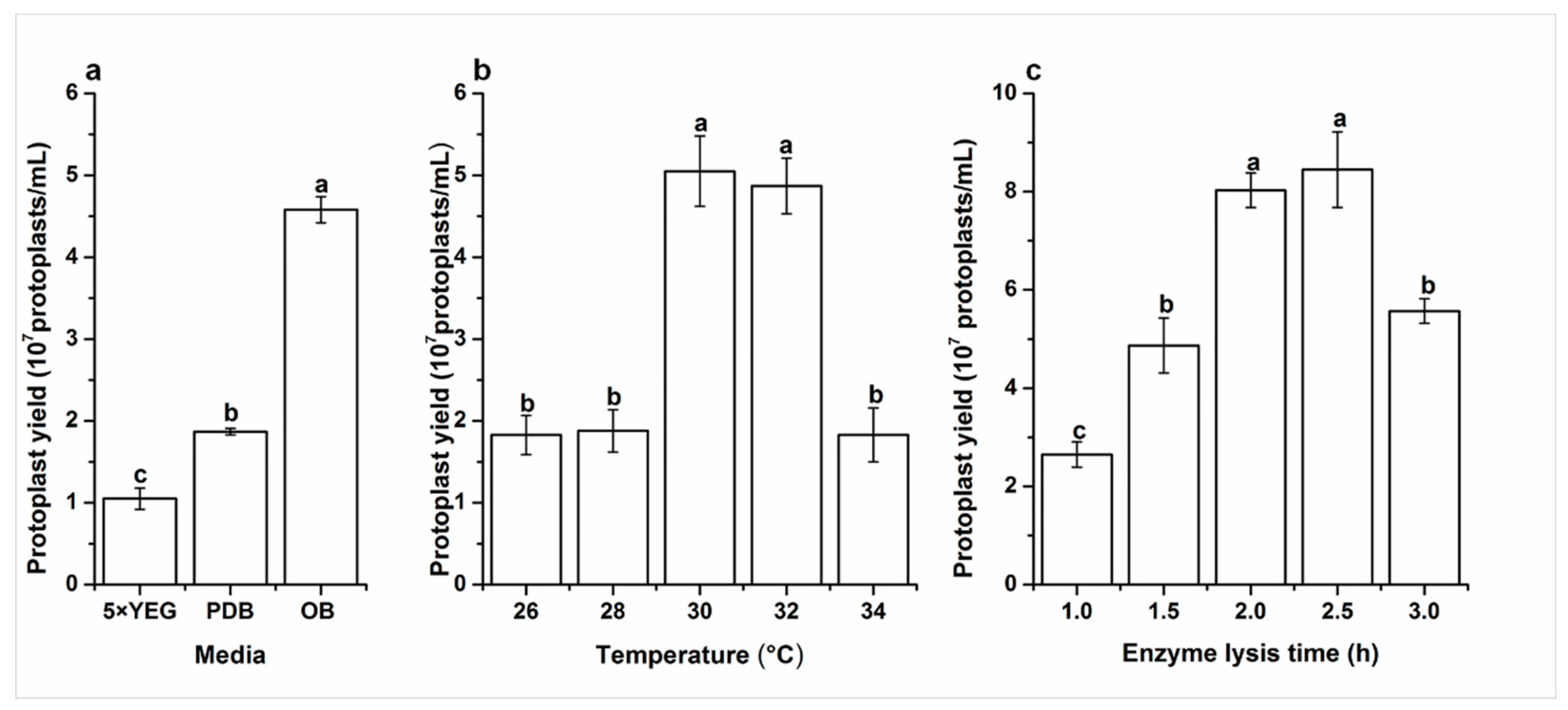
2.2. Effects of Media on Protoplast Yield
2.3. Effects of Digesting Temperature on Protoplast Yield
2.4. Effects of Enzyme Lysis Time on Protoplast Yield
2.5. Effects of Mycelia Weight on Protoplast Yield
2.6. Effects of Rotational Speed on Protoplast Isolation
2.7. Effects of Concentration of Lysing Enzyme on Protoplast Yield
2.8. Optimization of Production Conditions for Protoplasts by RSM
2.9. Transformation
2.10. Southern Blotting
3. Discussion
4. Materials and Methods
4.1. Fungal Strain, Media, and Reagents
4.2. Hygromycin Phosphotransferase Gene (hph) Fragment
4.3. Preparation of Protoplasts
4.4. Protoplast Yield Optimization
4.4.1. Effects of Enzyme on Protoplast Yield
4.4.2. Effects of Media on Protoplast Yield
4.4.3. Effects of Digesting Temperature on Protoplast Yield
4.4.4. Effects of Enzyme Lysis Time on Protoplast Yield
4.4.5. Effects of Mycelia Weight on Protoplast Yield
4.4.6. Effects of Rotational Speed on Protoplast Isolation
4.4.7. Effects of Concentration of Lysing Enzyme on Protoplast Yield
4.5. Response Surface Optimization Designs
4.6. Transformation
4.7. Southern Blotting
4.8. Data Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hernández-Restrepo, M.; Groenewald, J.Z.; Elliott, M.L.; Canning, G.; McMillan, V.E.; Crous, P.W. Take-all or nothing. Stud. Mycol. 2016, 83, 19–48. [Google Scholar] [CrossRef] [PubMed]
- Monsálvez, M.; Zapata, N.; Vargas, M.; Berti, M.; Bittner, M.; Hernández, V. Antifungal effects of n-hexane extract and essential oil of Drimys winteri bark against Take-All disease. Ind. Crops Prod. 2010, 31, 239–244. [Google Scholar] [CrossRef]
- Keenan, S.; Cromey, M.G.; Harrow, S.A.; Bithell, S.L.; Butler, R.C.; Beard, S.S.; Pitman, A.R. Quantitative PCR to detect Gaeumannomyces graminis var. tritici in symptomatic and non-symptomatic wheat roots. Australas. Plant Pathol. 2015, 44, 1–7. [Google Scholar] [CrossRef]
- Schesser, K.; Luder, A.; Henson, J.M. Use of polymerase chain reaction to detect the take-all fungus, Gaeumannomyces graminis, in infected wheat plants. Appl. Environ. Microbiol. 1991, 57, 553–556. [Google Scholar] [PubMed]
- Mcmillan, V.E.; Hammond-Kosack, K.E.; Gutteridge, R.J. Evidence that wheat cultivars differ in their ability to build up inoculum of the take-all fungus, Gaeumannomyces graminis var. tritici, under a first wheat crop. Plant Pathol. 2015, 60, 200–206. [Google Scholar] [CrossRef]
- Gutteridge, R.J.; Bateman, G.L.; Todd, A.D. Variation in the effects of take-all disease on grain yield and quality of winter cereals in field experiments. Pest Manag. Sci. 2003, 59, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Mcmillan, V.E.; Gutteridge, R.J.; Hammond-Kosack, K.E. Identifying variation in resistance to the take-all fungus, Gaeumannomyces graminis var. tritici, between different ancestral and modern wheat species. BMC Plant Biol. 2014, 14, 212. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Xie, L.; Xue, B.; Goodwin, P.H.; Quan, X.; Zheng, C.; Liu, T.; Lei, Z.; Yang, X.; Chao, Y.; et al. Comparative Transcriptome Profiling of the Early Infection of Wheat Roots by Gaeumannomyces graminis var. tritici. PLoS ONE 2015, 10, e0120691. [Google Scholar] [CrossRef] [PubMed]
- Travisi, C.M.; Nijkamp, P. Valuing environmental and health risk in agriculture: A choice experiment approach to pesticides in Italy. Ecol. Econ. 2008, 67, 598–607. [Google Scholar] [CrossRef]
- Vijver, M.G.; Hunting, E.R.; Nederstigt, T.A.; Tamis, W.L.; van den Brink, P.J.; van Bodegom, P.M. Postregistration monitoring of pesticides is urgently required to protect ecosystems. Environ. Toxicol. Chem. 2017, 36, 860–865. [Google Scholar] [CrossRef] [PubMed]
- Fincham, J.R. Transformation in fungi. Microbiol. Rev. 1989, 53, 148–170. [Google Scholar] [PubMed]
- Robinson, M.; Sharon, A. Transformation of the bioherbicide Colletotrichum gloeosporioides f. sp. aeschynomene by electroporation of germinated conidia. Curr. Genet. 1999, 36, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Irie, T.; Honda, Y.; Watanabe, T.; Kuwahara, M. Efficient transformation of filamentous fungus Pleurotus ostreatus using single-stranded carrier DNA. Appl. Microbiol. Biotechnol. 2001, 55, 563–565. [Google Scholar] [CrossRef] [PubMed]
- Stanway, C.; Buck, K.W. Infection of Protoplasts of the Wheat Take-all Fungus, Gaeumannomyces graminis var. tritici, with Double-stranded RNA Viruses. J. Gen. Virol. 1984, 65, 2061–2065. [Google Scholar] [CrossRef]
- Henson, J.M.; Blake, N.K.; Pilgeram, A.L. Transformation of Gaeumannomyces graminis to benomyl resistance. Curr. Genet. 1988, 14, 113–117. [Google Scholar] [CrossRef]
- Park, J.; Radwan, O.; Martin, B.; Wilkinson, H.T.; Fouly, H.M. Transformation of Gaeumannomyces graminis with beta-Glucuronidase Reporter and Hygromycin Resistance Genes. J. Phytopathol. 2011, 159, 317–320. [Google Scholar] [CrossRef]
- Bowyer, P.; Musker, R.; Osbourn, A.E.; Clarke, B.; Caten, C.; Daniels, M.J. Production of mutants of Gaeumannomyces graminis var. tritici and var. avenae by 4-nitroquinolene-oxide treatment of protoplasts. Fungal Genet. Rep. 1992, 39, 13–14. [Google Scholar] [CrossRef]
- Zhou, W.W.; He, Y.L.; Niu, T.G.; Zhong, J.J. Optimization of fermentation conditions for production of anti-TMV extracellular ribonuclease by Bacillus cereus using response surface methodology. Bioprocess Biosyst. Eng. 2010, 33, 657–663. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; You, Q.; Jiang, Z. Optimization of enzyme assisted extraction of polysaccharides from Tricholoma matsutake by response surface methodology. Carbohydr. Polym. 2011, 86, 1358–1364. [Google Scholar] [CrossRef]
- Hu, G.P.; Zhao, Y.; Song, F.Q.; Liu, B.; Vasseur, L.; Douglas, C.; You, M.S. Isolation, identification and cyfluthrin-degrading potential of a novel Lysinibacillus sphaericus strain, FLQ-11-1. Res. Microbiol. 2014, 165, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Khairudin, N.; Basri, M.; Fard, H.M.; Samson, S.; Ashari, S.E. Enhancing the bioconversion of azelaic acid to its derivatives by response surface methodology. Molecules 2018, 23, 397. [Google Scholar] [CrossRef] [PubMed]
- Altemimi, A.; Lightfoot, D.A.; Kinsel, M.; Watson, D.G. Employing response surface methodology for the optimization of ultrasound assisted extraction of lutein and β-carotene from spinach. Molecules 2015, 20, 6611–6625. [Google Scholar] [CrossRef] [PubMed]
- Muntari, B.; Amid, A.; Mel, M.; Jami, M.S.; Salleh, H.M. Recombinant bromelain production in Escherichia coli: Process optimization in shake flask culture by response surface methodology. AMB Express 2012, 2, 12. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Sun, Y.B.; He, H.W.; Feng, J.T.; Zhang, X.; Han, L.R. Optimization of medium compositions to improve a novel glycoprotein production by Streptomyces kanasenisi ZX01. AMB Express 2017, 7, 6. [Google Scholar] [CrossRef] [PubMed]
- Mishra, N.C.; Tatum, E.L. Non-Mendelian inheritance of DNA-induced inositol independence in Neurospora. Proc. Natl. Acad. Sci. USA 1973, 70, 3875–3879. [Google Scholar] [CrossRef] [PubMed]
- Ge, C.Y.; Duan, Y.B.; Zhou, M.G.; Chen, C.J. A protoplast transformation system for gene deletion and complementation in Sclerotinia sclerotiorum. J. Phytopathol. 2013, 161, 800–806. [Google Scholar] [CrossRef]
- Zhang, Y.J.; Xie, M.; Zhang, X.L.; Peng, D.L.; Yu, W.B.; Li, Q.; Li, Q.; Zhao, J.J.; Zhang, Z.R. Establishment of polyethylene-glycol-mediated protoplast transformation for Lecanicillium lecanii and development of virulence-enhanced strains against Aphis gossypii. Pest Manag. Sci. 2016, 72, 1951–1958. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, K.T.; Ho, Q.N.; Do, L.T.B.X.; Mai, L.T.D.; Pham, D.N.; Tran, H.T.T.; Le, D.H.; Nguyen, H.Q.; Tran, V.T. A new and efficient approach for construction of uridine/uracil auxotrophic mutants in the filamentous fungus Aspergillus oryzae using Agrobacterium tumefaciens-mediated transformation. World J. Microbiol. Biotechnol. 2017, 33, 107. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.P.; Gao, T.; Zheng, Z.T.; Chen, C.J.; Zhou, M.G. An economic and highly efficient system of protoplast transformation for Fusarium graminearum. Mycosystema 2013, 32, 891–898. [Google Scholar]
- Zhang, C.; Zong, H.; Zhuge, B.; Lu, X.; Fang, H.; Zhu, J.; Zhuge, J. Protoplast preparation and polyethylene glycol (PEG)-mediated transformation of Candida glycerinogenes. Biotechnol. Bioprocess Eng. 2016, 21, 95–102. [Google Scholar] [CrossRef]
- Maier, F.J.; Malz, S.; Lösch, A.P.; Lacour, T.; Schäfer, W. Development of a highly efficient gene targeting system for Fusarium graminearum using the disruption of a polyketide synthase gene as a visible marker. FEMS Yeast Res. 2005, 5, 653–662. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, D.G. Gene targeting in Physcomitrella patens. Curr. Opin. Plant Biol. 2001, 4, 143–150. [Google Scholar] [CrossRef]
- Wang, M.; Wang, L.; Han, L.; Zhang, X.; Feng, J. The effect of carabrone on mitochondrial respiratory chain complexes in Gaeumannomyces graminis. J. Appl. Microbiol. 2017, 123, 1100–1110. [Google Scholar] [CrossRef] [PubMed]
- Nakayashiki, H.; Hanada, S.; Quoc, N.B.; Kadotani, N.; Tosa, Y.; Mayama, S. RNA silencing as a tool for exploring gene function in ascomycete fungi. Fungal Genet. Biol. 2005, 42, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Gao, Q.; Huang, M.; Liu, Y.; Liu, Z.; Liu, X.; Ma, Z. Characterization of RNA silencing components in the plant pathogenic fungus Fusarium graminearum. Sci. Rep. 2015, 5, 12500. [Google Scholar] [CrossRef] [PubMed]
- Tetorya, M.; Rajam, M.V. RNA silencing of PEX6 gene causes decrease in pigmentation, sporulation and pathogenicity of Fusarium oxysporum. Plant Pathol. 2017, 67. [Google Scholar] [CrossRef]
- Jia, X.; Zhang, X.; Qu, J.; Han, R. Optimization conditions of wheat mesophyll protoplast isolation. Agric. Sci. 2016, 7, 850–858. [Google Scholar] [CrossRef]
- Cheng, W.; Song, X.S.; Li, H.P.; Cao, L.H.; Sun, K.; Qiu, X.L.; Xu, Y.B.; Yang, P.; Huang, T.; Zhang, J.B.; et al. Host-induced gene silencing of an essential chitin synthase gene confers durable resistance to Fusarium head blight and seedling blight in wheat. Plant Biotechnol. J. 2015, 13, 1335–1345. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds Gaeumannomyces tritici (number ACCC30310) are available from the authors. |


| Treatment 1 | Protoplast Yield (107 Protoplasts/mL) |
|---|---|
| 0.25% L | 2.07 ± 0.02b |
| 0.25% D | 0.17 ± 0.02d |
| 0.25% S | 0.03 ± 0.02d |
| 0.25% S + 0.25% L | 2.10 ± 0.28ab |
| 0.25% S + 0.25% D | 1.38 ± 0.17c |
| 0.25% L + 0.25% D | 2.37 ± 0.27ab |
| 0.25% S + 0.25% D + 0.25% L | 2.68 ± 0.24a |
| Run | X1 | X2 | X3 | Experimental Value (107 Protoplasts/mL) a | Predicted Value (107 Protoplasts/mL) | Residual |
|---|---|---|---|---|---|---|
| 1 | 0 | −1 | −1 | 8.05 | 8.07 | −0.02 |
| 2 | −1 | 0 | −1 | 5.69 | 5.77 | −0.08 |
| 3 | 1 | −1 | 0 | 9.14 | 9.41 | −0.27 |
| 4 | 0 | 0 | 0 | 9.33 | 9.37 | −0.04 |
| 5 | 1 | 0 | −1 | 8.21 | 7.93 | 0.28 |
| 6 | 1 | 0 | 1 | 8.69 | 8.59 | 0.10 |
| 7 | −1 | −1 | 0 | 5.97 | 5.85 | 0.12 |
| 8 | −1 | 0 | 1 | 5.43 | 5.71 | −0.28 |
| 9 | 0 | −1 | 1 | 8.76 | 8.57 | 0.19 |
| 10 | 1 | 1 | 0 | 8.02 | 8.13 | −0.11 |
| 11 | 0 | 0 | 0 | 9.23 | 9.37 | −0.14 |
| 12 | 0 | 0 | 0 | 9.56 | 9.37 | 0.19 |
| 13 | 0 | 1 | −1 | 7.84 | 8.03 | −0.19 |
| 14 | 0 | 1 | 1 | 8.15 | 8.13 | 0.02 |
| 15 | −1 | 1 | 0 | 6.92 | 6.65 | 0.27 |
| Source | Sum of Squares | DF a | Mean Square | F Value | p-Value Prob > F |
|---|---|---|---|---|---|
| Model | 24.85 | 9 | 2.76 | 29.06 | 0.0009 |
| X1 | 12.63 | 1 | 12.63 | 132.87 | <0.0001 |
| X2 | 0.12 | 1 | 0.12 | 1.29 | 0.3076 |
| X3 | 0.19 | 1 | 0.19 | 2.02 | 0.2142 |
| X1X2 | 1.07 | 1 | 1.07 | 11.27 | 0.0202 |
| X1X3 | 0.14 | 1 | 0.14 | 1.44 | 0.2838 |
| X2X3 | 0.04 | 1 | 0.04 | 0.42 | 0.5451 |
| X12 | 8.62 | 1 | 8.62 | 90.72 | 0.0002 |
| X22 | 0.41 | 1 | 0.41 | 4.31 | 0.0926 |
| X32 | 2.61 | 1 | 2.61 | 27.45 | 0.0034 |
| Residual | 0.48 | 5 | 0.095 | ||
| Lack of Fit | 0.42 | 3 | 0.14 | 4.86 | 0.1752 |
| Pure Error | 0.057 | 2 | 0.029 | ||
| Total | 25.32 | 14 |
| Amount of Transform DNA (µg) | Amount of Transformants | Transformation Efficiency (Transformants/µg DNA) |
|---|---|---|
| 4 | 185.00 ± 5.51 | 46.25 ± 1.38b |
| 6 | 324.00 ± 3.61 | 54.00 ± 0.60a |
| 8 | 391.33 ± 4.33 | 48.92 ± 0.54b |
| Symbols | Independent Variables | −1 | 0 | 1 |
|---|---|---|---|---|
| X1 | Temperature (°C) | 28 | 30 | 32 |
| X2 | Enzyme lysis time (h) | 2 | 2.5 | 3 |
| X3 | Lysing enzyme concentration (%) | 0.5 | 1 | 2 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, M.; Zhang, J.; Wang, L.; Han, L.; Zhang, X.; Feng, J. Optimization of Production Conditions for Protoplasts and Polyethylene Glycol-Mediated Transformation of Gaeumannomyces tritici. Molecules 2018, 23, 1253. https://doi.org/10.3390/molecules23061253
Wang M, Zhang J, Wang L, Han L, Zhang X, Feng J. Optimization of Production Conditions for Protoplasts and Polyethylene Glycol-Mediated Transformation of Gaeumannomyces tritici. Molecules. 2018; 23(6):1253. https://doi.org/10.3390/molecules23061253
Chicago/Turabian StyleWang, Mei, Jie Zhang, Lanying Wang, Lirong Han, Xing Zhang, and Juntao Feng. 2018. "Optimization of Production Conditions for Protoplasts and Polyethylene Glycol-Mediated Transformation of Gaeumannomyces tritici" Molecules 23, no. 6: 1253. https://doi.org/10.3390/molecules23061253
APA StyleWang, M., Zhang, J., Wang, L., Han, L., Zhang, X., & Feng, J. (2018). Optimization of Production Conditions for Protoplasts and Polyethylene Glycol-Mediated Transformation of Gaeumannomyces tritici. Molecules, 23(6), 1253. https://doi.org/10.3390/molecules23061253