Design, Synthesis, and Docking Studies of New Torin2 Analogs as Potential ATR/mTOR Kinase Inhibitors †
Abstract
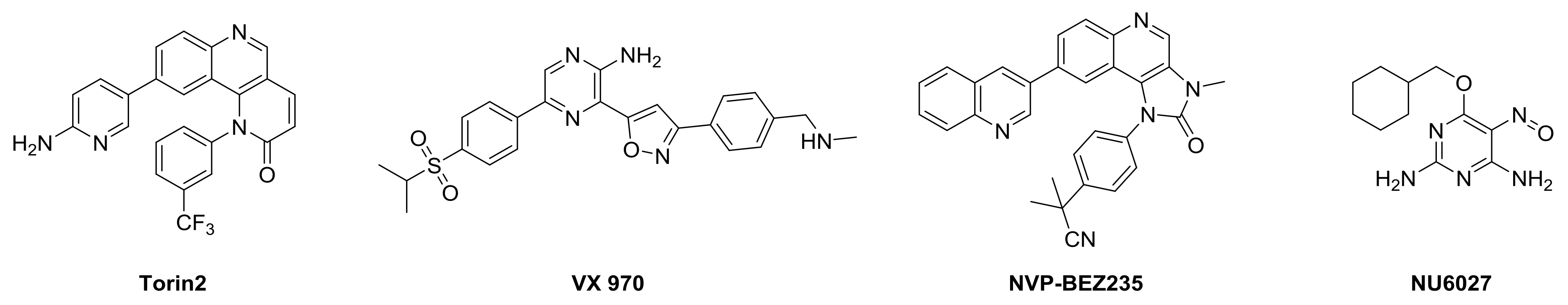
:1. Introduction
2. Results
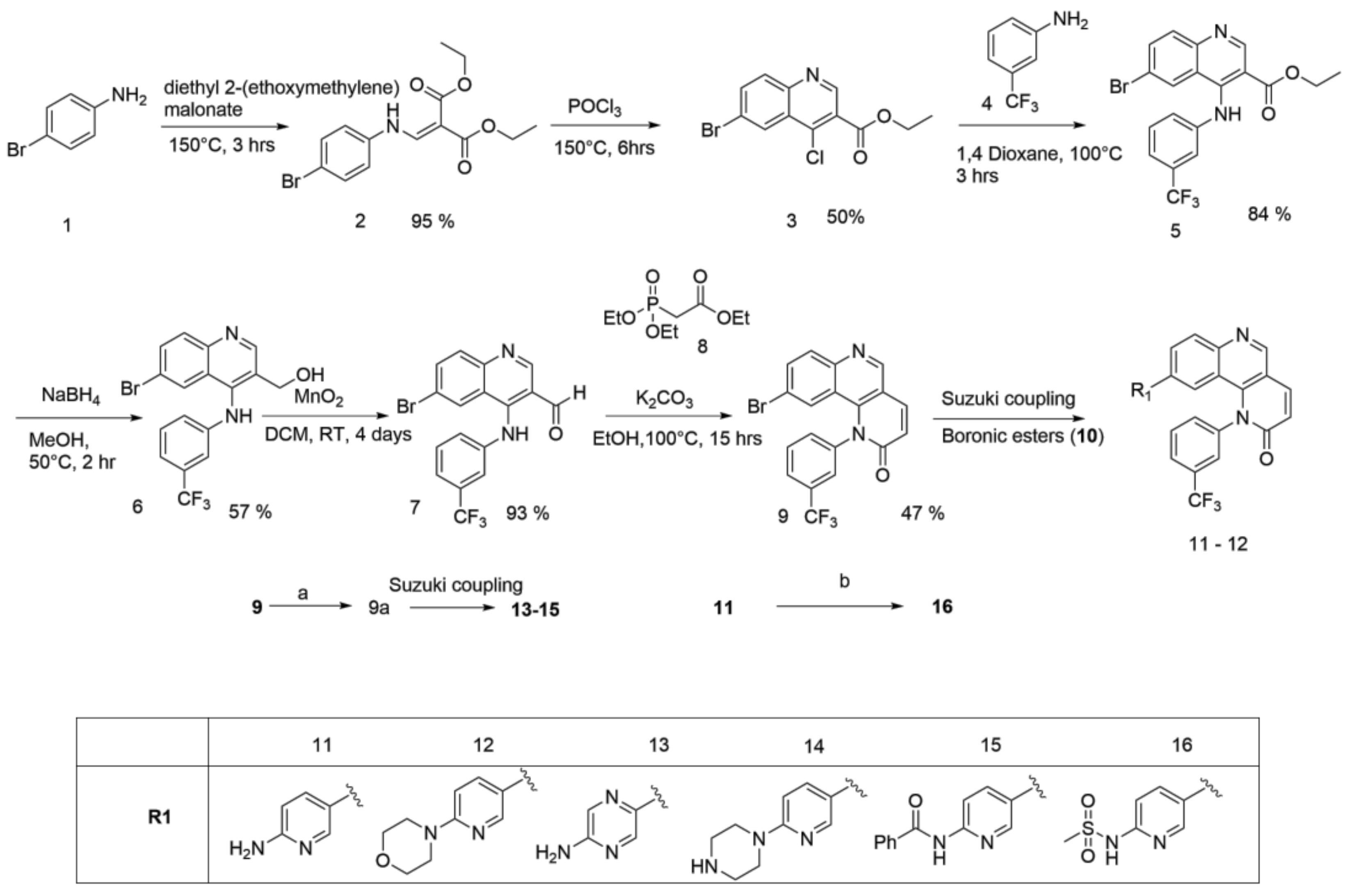
2.1. Synthesis
2.1.1. Scheme 1
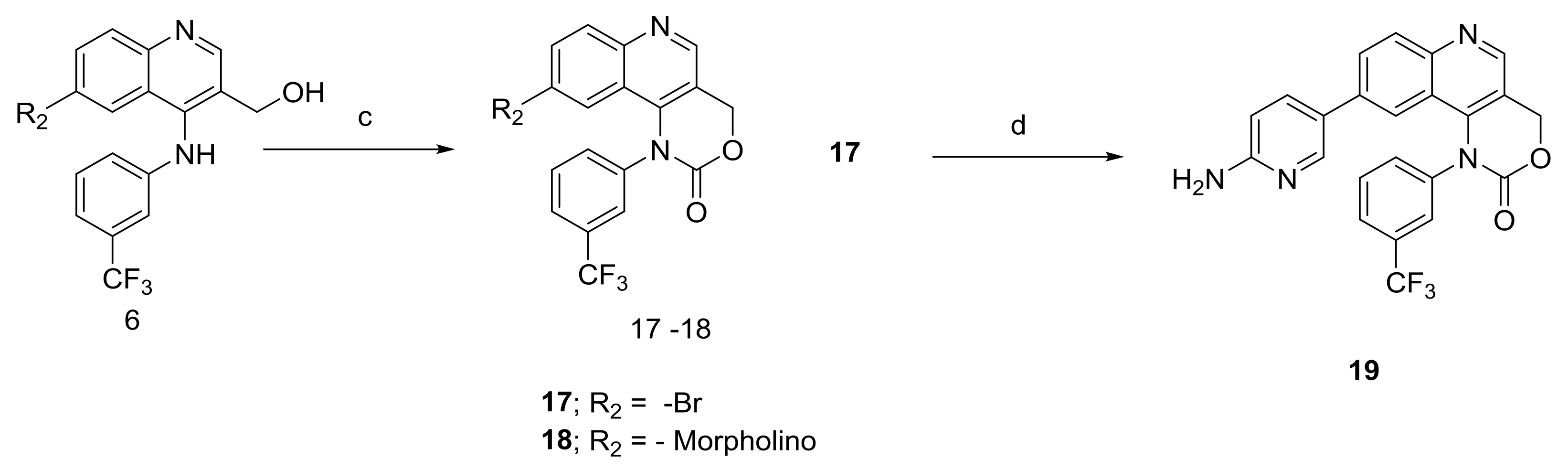
2.1.2. Scheme 2
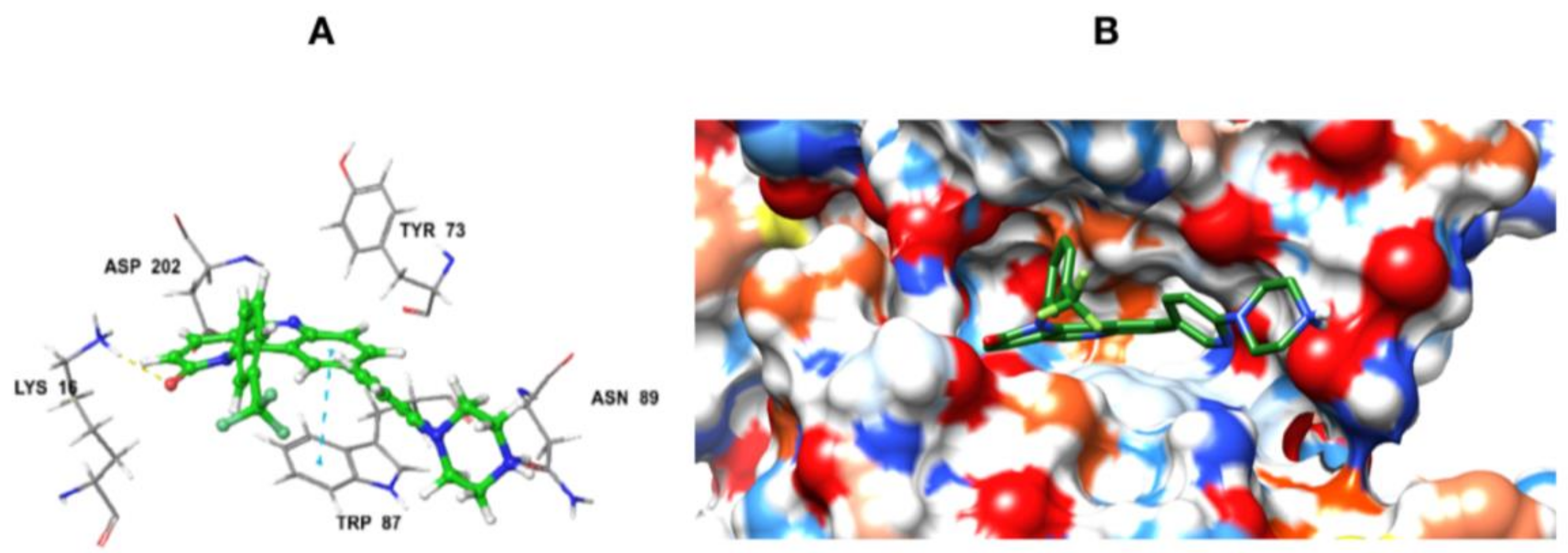
2.2. Homology Modeling and Docking Studiess
2.2.1. Homology Modeling
2.2.2. Quality Assessment/Model Validation
2.2.3. Molecular Dynamic Simulations and Docking Studies with Torin2
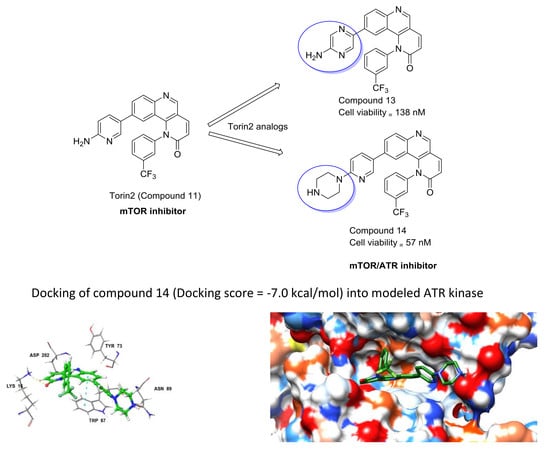
2.2.4. Docking of Designed Inhibitors and Binding Pose Analysis
2.3. Cell Viability Assay
2.4. Immunoblot Assay
3. Discussion
4. Materials and Methods
4.1. Chemistry
4.1.1. Synthesis of Torin2 Analogs (Compounds 11–12)


4.1.2. Synthesis of 9-(5-Aminopyrazin-2-yl)-1-(3-(trifluoromethyl)phenyl)benzo[h][1,6]naphtha yridin-2(1H)-one (Compound 13)

4.1.3. Synthesis of 9-(6-(Piperazin-1-yl)pyridin-3-yl)-1-(3-(trifluoromethyl)phenyl)benzo[h][1,6]naphthyridin-2(1H)-one (compound 14)

4.1.4. Synthesis of N-(5-(2-Oxo-1-(3-(trifluoromethyl)phenyl)-1,2-dihydrobenzo[h][1,6]naphthyridin-9-yl)pyridin-2-yl)benzamide (compound 15)

4.1.5. Synthesis of N-(5-(2-Oxo-1-(3-(trifluoromethyl)phenyl)-1,2-dihydrobenzo[h][1,6] naphthyridin-9-yl)pyridin-2-yl)methanesulfonamide (Compound 16)

4.1.6. Synthesis of 9-Bromo-1-(3-(trifluoromethyl)phenyl)-1,4-dihydro-2H-[1,3]oxazino[5,4-c]quinolin-2-one and 9-morpholino-1-(3-(trifluoromethyl)phenyl)-1,4-dihydro-2H-[1,3]oxazino[5,4-c]quinolin-2-one (Compound 17–18)


4.1.7. Synthesis of 9-(6-Aminopyridin-3-yl)-1-(3-(trifluoromethyl)phenyl)-1,4-dihydro-2H-[1,3]oxazino[5,4-c]quinolin-2-one (Compound 19)

4.2. In Silco Studies
4.2.1. Homology Modeling of ATR Kinase
4.2.2. Model Validation
4.2.3. MD Simulations
4.2.4. Docking Studies
4.3. Cell Based Studies
Materials and Methods
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Fokas, E.; Prevo, R.; Hammond, E.M.; Brunner, T.B.; McKenna, W.G.; Muschel, R.J. Targeting ATR in DNA damage response and cancer therapeutics. Cancer Treat. Rev. 2014, 40, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.P.; Bartek, J. The DNA-damage response in human biology and disease. Nature 2009, 461, 1071–1078. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Xu, Z.P.; Huang, Y.; Hamrick, H.E.; Duerksen-Hughes, P.J.; Yu, Y.N. ATM and ATR: Sensing DNA damage. World J. Gastroenterol. 2004, 10, 155–160. [Google Scholar] [CrossRef] [PubMed]
- Llona-Minguez, S.; Hoglund, A.; Jacques, S.A.; Koolmeister, T.; Helleday, T. Chemical strategies for development of ATR inhibitors. Expert Rev. Mol. Med. 2014, 9, 10. [Google Scholar] [CrossRef] [PubMed]
- Wagner, J.M.; Kaufmann, S.H. Prospects for the Use of ATR Inhibitors to Treat Cancer. Pharmaceuticals 2010, 3, 1311–1334. [Google Scholar] [CrossRef] [PubMed]
- Charrier, J.D.; Durrant, S.J.; Golec, J.M.; Kay, D.P.; Knegtel, R.M.; MacCormick, S.; Mortimore, M.; O’Donnell, M.E.; Pinder, J.L.; Reaper, P.M.; et al. Discovery of potent and selective inhibitors of ataxia telangiectasia mutated and Rad3 related (ATR) protein kinase as potential anticancer agents. J. Med. Chem. 2011, 54, 2320–2330. [Google Scholar] [CrossRef] [PubMed]
- Hall, A.B.; Newsome, D.; Wang, Y.; Boucher, D.M.; Eustace, B.; Gu, Y.; Hare, B.; Johnson, M.A.; Milton, S.; Murphy, C.E.; et al. Potentiation of Tumor Responses to DNA Damaging Therapy by the Selective ATR Inhibitor VX-970. Oncotarget 2014, 5, 5674–5685. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Wang, J.; Kang, S.A.; Thoreen, C.C.; Hur, W.; Ahmed, T.; Sabatini, D.M.; Gray, N.S. Discovery of 9-(6-Aminopyridin-3-yl)-1-(3-(trifluoromethyl)phenyl)benzo[h][1,6]naphthyridin-2(1H)-one (Torin2) as a Potent, Selective, and Orally Available Mammalian Target of Rapamycin (mTOR) Inhibitor for Treatment of Cancer. J. Med. Chem. 2011, 54, 1473–1480. [Google Scholar] [CrossRef] [PubMed]
- Toledo, L.I.; Murga, M.; Zur, R.; Soria, R.; Rodriguez, A.; Martinez, S.; Oyarzabal, J.; Pastor, J.; Bischoff, J.R.; Fernandez-Capetillo, O. A cell-based screen identifies ATR inhibitors with synthetic lethal properties for cancer-associated mutations. Nat. Struct. Mol. Biol. 2011, 18, 721–727. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Xu, C.; Kirubakaran, S.; Zhang, X.; Hur, W.; Liu, Y.; Kwiatkowski, N.P.; Wang, J.; Westover, K.D.; Gao, P.; et al. Characterization of Torin2, an ATP-competitive inhibitor of mTOR, ATM, and ATR. Cancer Res. 2013, 73, 2574–2586. [Google Scholar] [CrossRef] [PubMed]
- Malm, J.; Ringom, R.; Caldirola, P.; Westman, J. Substituted Quinolines for Use as Vegf Inhibitors. Google Patents No. WO2010133669A1, 20 May 2010. [Google Scholar]
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T.; Kiefer, F.; Cassarino, T.G.; Bertoni, M.; Bordoli, L.; et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014, 42, W252–W258. [Google Scholar] [CrossRef] [PubMed]
- Maiti, R.; Van Domselaar, G.H.; Zhang, H.; Wishart, D.S. SuperPose: A simple server for sophisticated structural superposition. Nucleic Acids Res. 2004, 32, W590–W594. [Google Scholar] [CrossRef] [PubMed]
- Zaytseva, Y.Y.; Valentino, J.D.; Gulhati, P.; Mark Evers, B. mTOR inhibitors in cancer therapy. Cancer Lett. 2012, 319, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Remmert, M.; Biegert, A.; Hauser, A.; Soding, J. HHblits: Lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat. Methods 2011, 9, 173–175. [Google Scholar] [CrossRef] [PubMed]
- Guex, N.; Peitsch, M.C. SWISS-MODEL and the Swiss-Pdb Viewer: An environment for comparative protein modeling. Electrophoresis 1997, 18, 2714–2723. [Google Scholar] [CrossRef] [PubMed]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra Precision Glide: Docking and Scoring Incorporating a Model of Hydrophobic Enclosure for Protein-Ligand Complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Wiederstein, M.; Sippl, M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef] [PubMed]
- Lovell, S.C.; Davis, I.W.; Arendall, W.B., 3rd; de Bakker, P.I.; Word, J.M.; Prisant, M.G.; Richardson, J.S.; Richardson, D.C. Structure validation by Calpha geometry: Phi,psi and Cbeta deviation. Proteins 2003, 50, 437–450. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Madhavi Sastry, G.; Adzhigirey, M.; Day, T.; Annabhimoju, R.; Sherman, W. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 2013, 27, 221–234. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds reported are available from the authors. |





| S.no | List of Compounds | Glide XP Score (kcal/mol) | Key Interacting Residues | Hydrogen Bonding Residues | Hydrogen Bonding Distance (Å) |
|---|---|---|---|---|---|
| 1 | Compound 11 (Torin2) | −8.0 | TRP 87(π-π stacking), LYS 16, ASN 89 | LYS 16, ASN 89 | 2.00–2.15 |
| 2 | Compound 12 | −7.2 | TRP 87(π-π stacking), LYS 16 | LYS 16 | 2.13 |
| 3 | Compound 13 | −8.1 | TRP 87(π-π stacking), LYS 16, ASN 89, VAL 88, ILE 189, LEU 190 | LYS 16, ASN 89, VAL88, ILE 189, LEU 190 | 2.10–2.95 |
| 4 | Compound 14 | −7.0 | TRP 87(π-π stacking), LYS 16, ASN 89 | LYS 16, ASN 89 | 2.31–2.87 |
| 5 | Compound 15 | −6.5 | TRP 87(π-π stacking), LYS 16, ASP 202 | LYS 16, ASP 202 | 2.07–1.80 |
| 6 | Compound 16 | −8.2 | TRP 87(π-π stacking), LYS 16, ASN 89 | LYS 16, ASN 89 | 2.05–2.25 |
| 7 | Compound 17 | −3.4 | TRP 87(π-π stacking), LYS 16 | LYS 16 | 2.27 |
| 8 | Compound 18 | −4.0 | TRP 87(π-π stacking), LYS 16 | LYS 16 | 2.24 |
| 9 | Compound 19 | −4.3 | TRP 87(π-π stacking), LYS 16, ASN 89 | LYS 16, ASN 89 | 2.35–2.26 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shaik, A.; Bhakuni, R.; Kirubakaran, S. Design, Synthesis, and Docking Studies of New Torin2 Analogs as Potential ATR/mTOR Kinase Inhibitors. Molecules 2018, 23, 992. https://doi.org/10.3390/molecules23050992
Shaik A, Bhakuni R, Kirubakaran S. Design, Synthesis, and Docking Studies of New Torin2 Analogs as Potential ATR/mTOR Kinase Inhibitors. Molecules. 2018; 23(5):992. https://doi.org/10.3390/molecules23050992
Chicago/Turabian StyleShaik, Althaf, Rashmi Bhakuni, and Sivapriya Kirubakaran. 2018. "Design, Synthesis, and Docking Studies of New Torin2 Analogs as Potential ATR/mTOR Kinase Inhibitors" Molecules 23, no. 5: 992. https://doi.org/10.3390/molecules23050992