A Comprehensive Genome Survey Provides Novel Insights into Bile Salt Hydrolase (BSH) in Lactobacillaceae
Abstract
:1. Introduction
2. Results
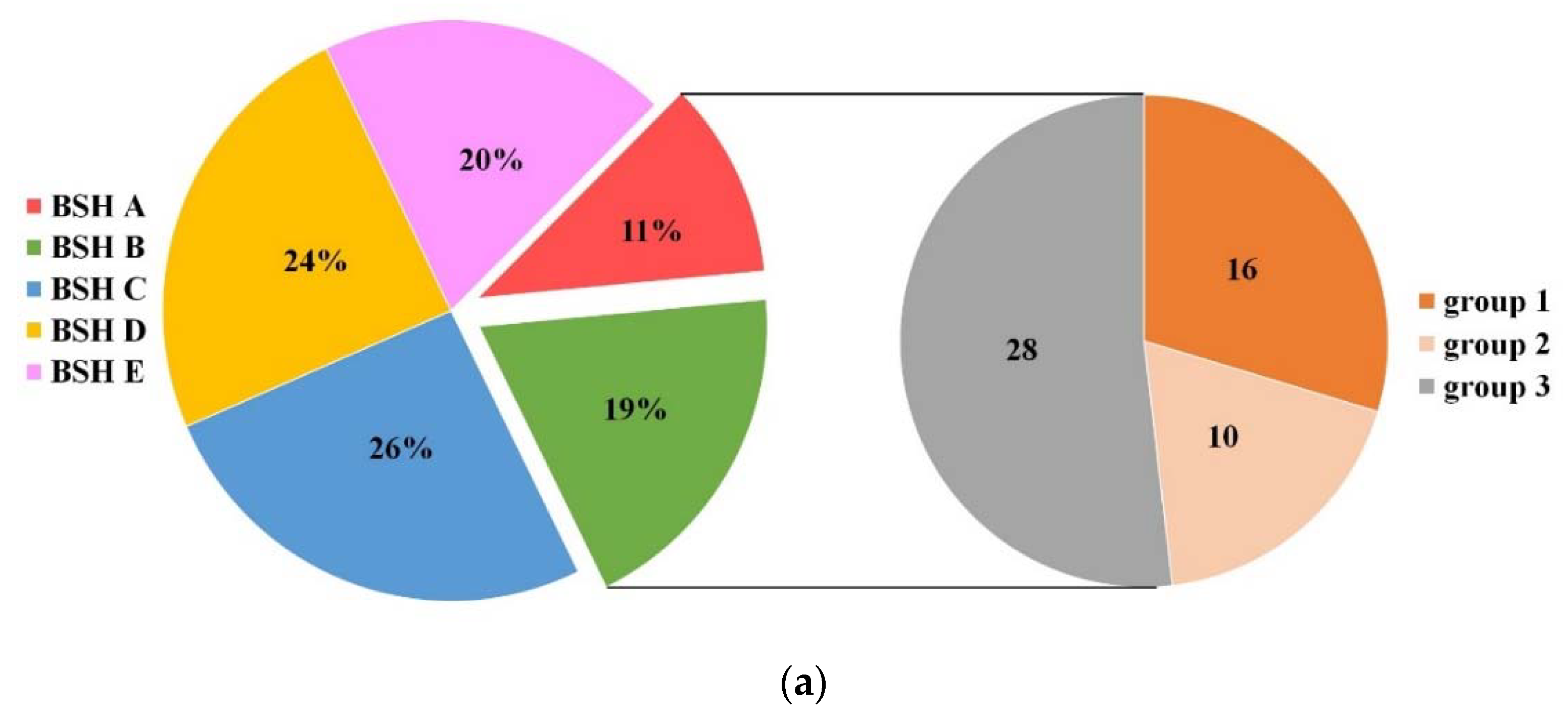
2.1. Establishment of BSH Database in Lactobacillaceae
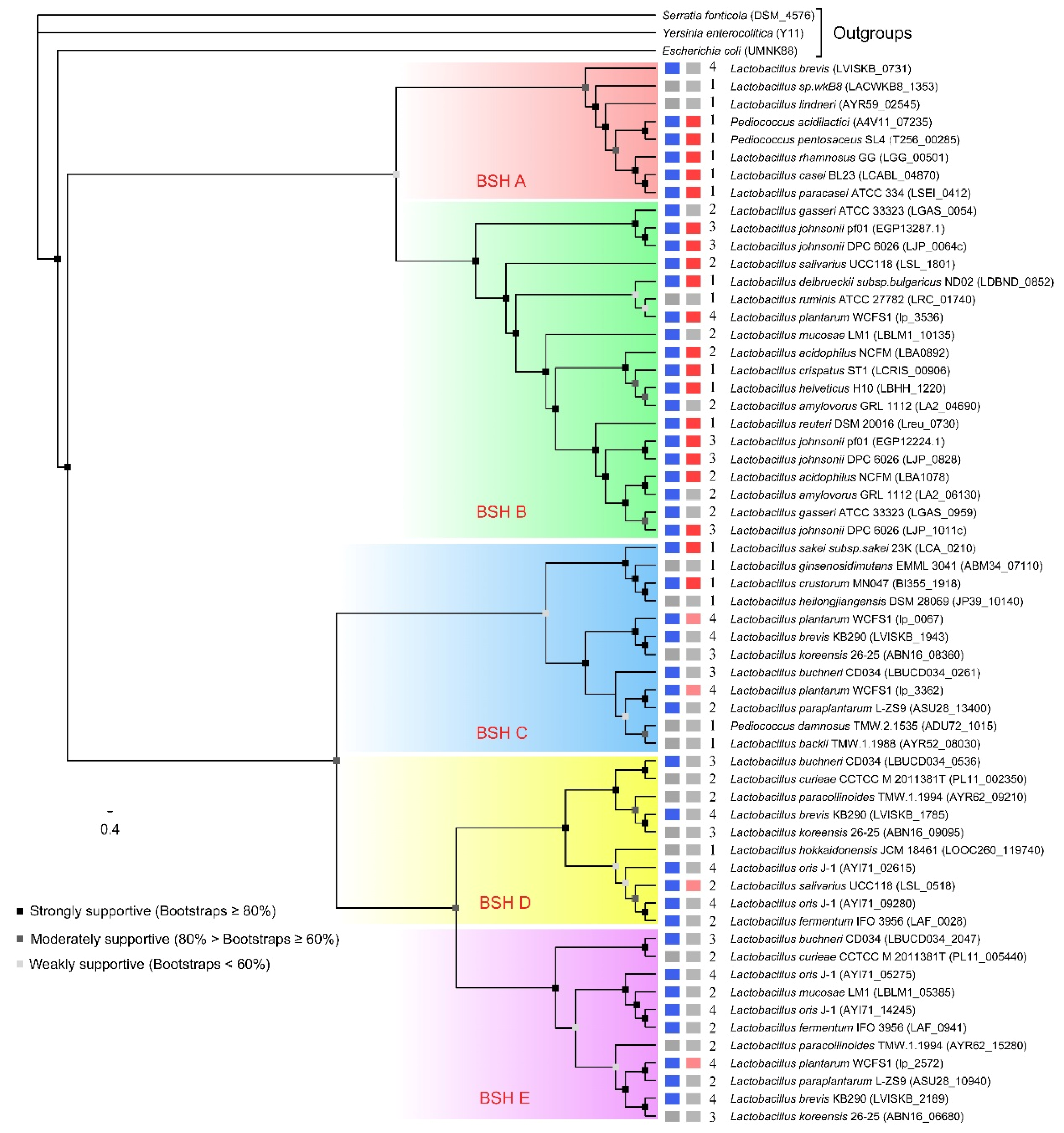
2.2. Phylogenetic Analysis
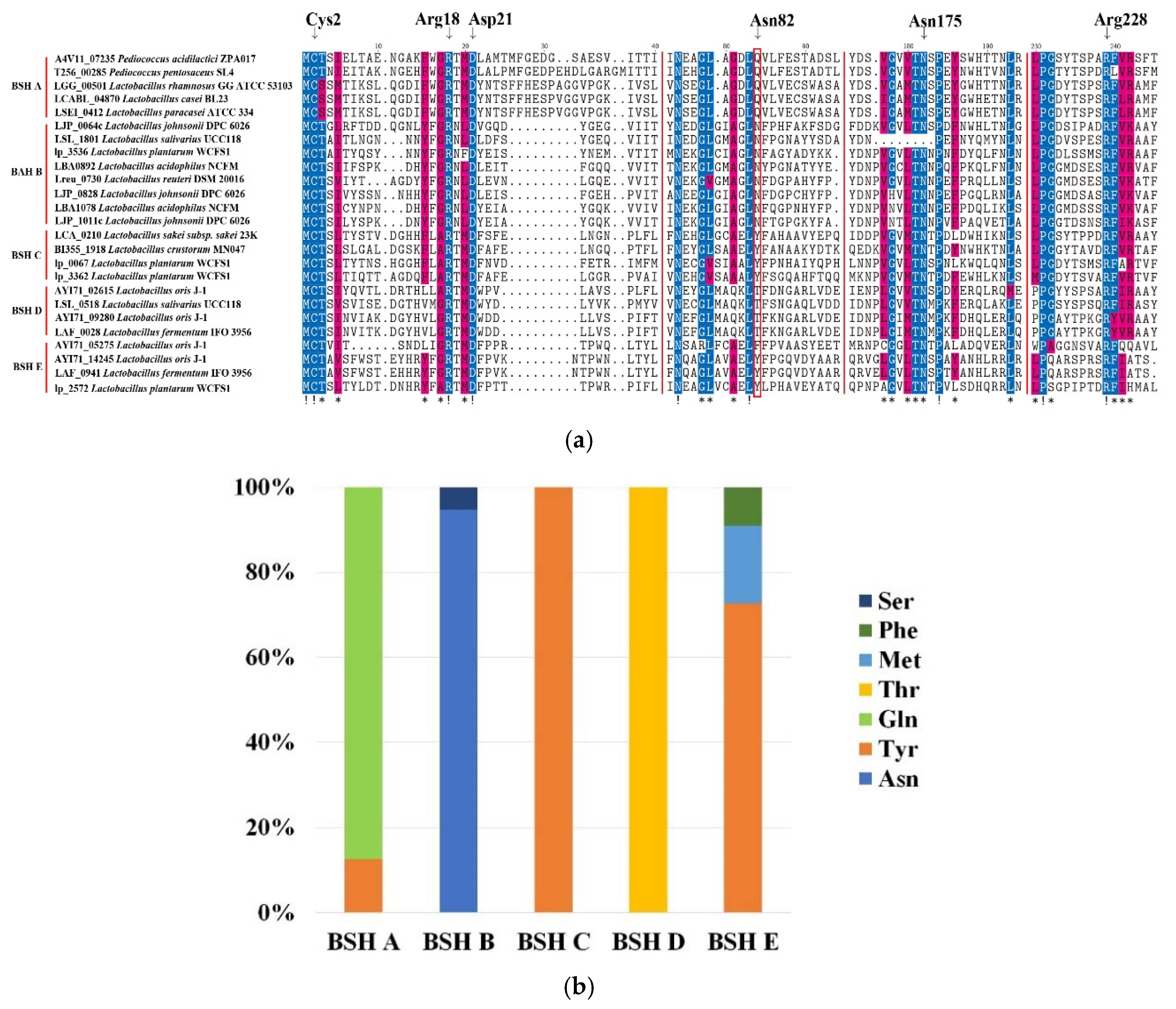
2.3. Sequence Analysis of BSH
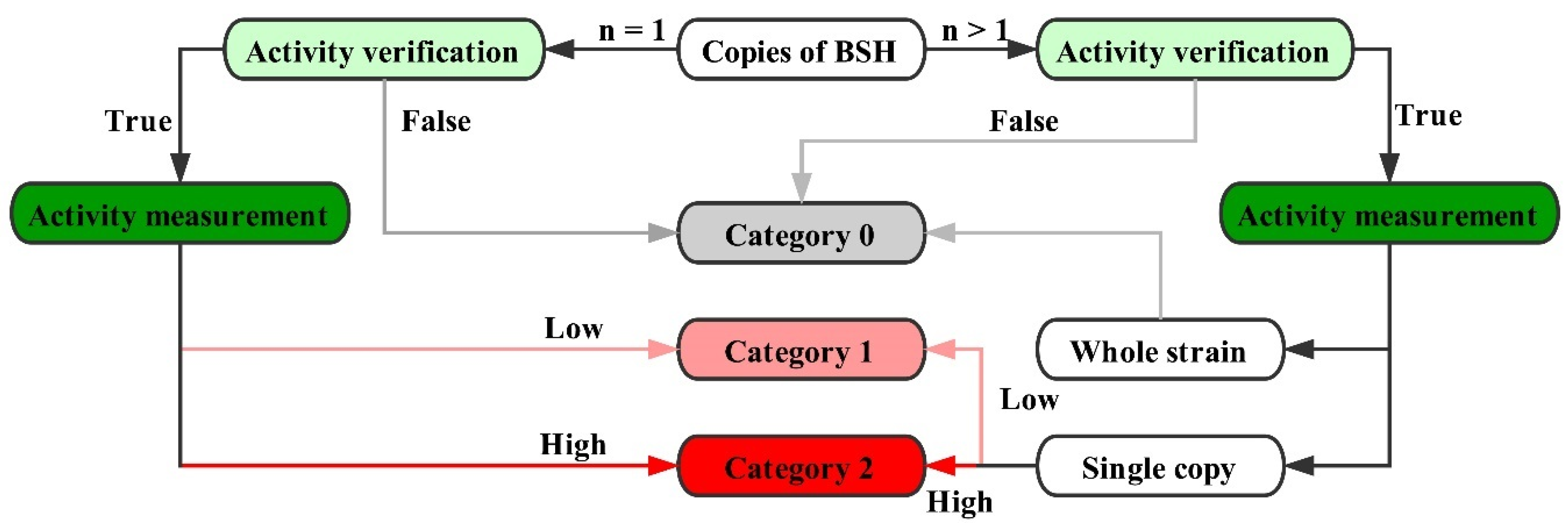
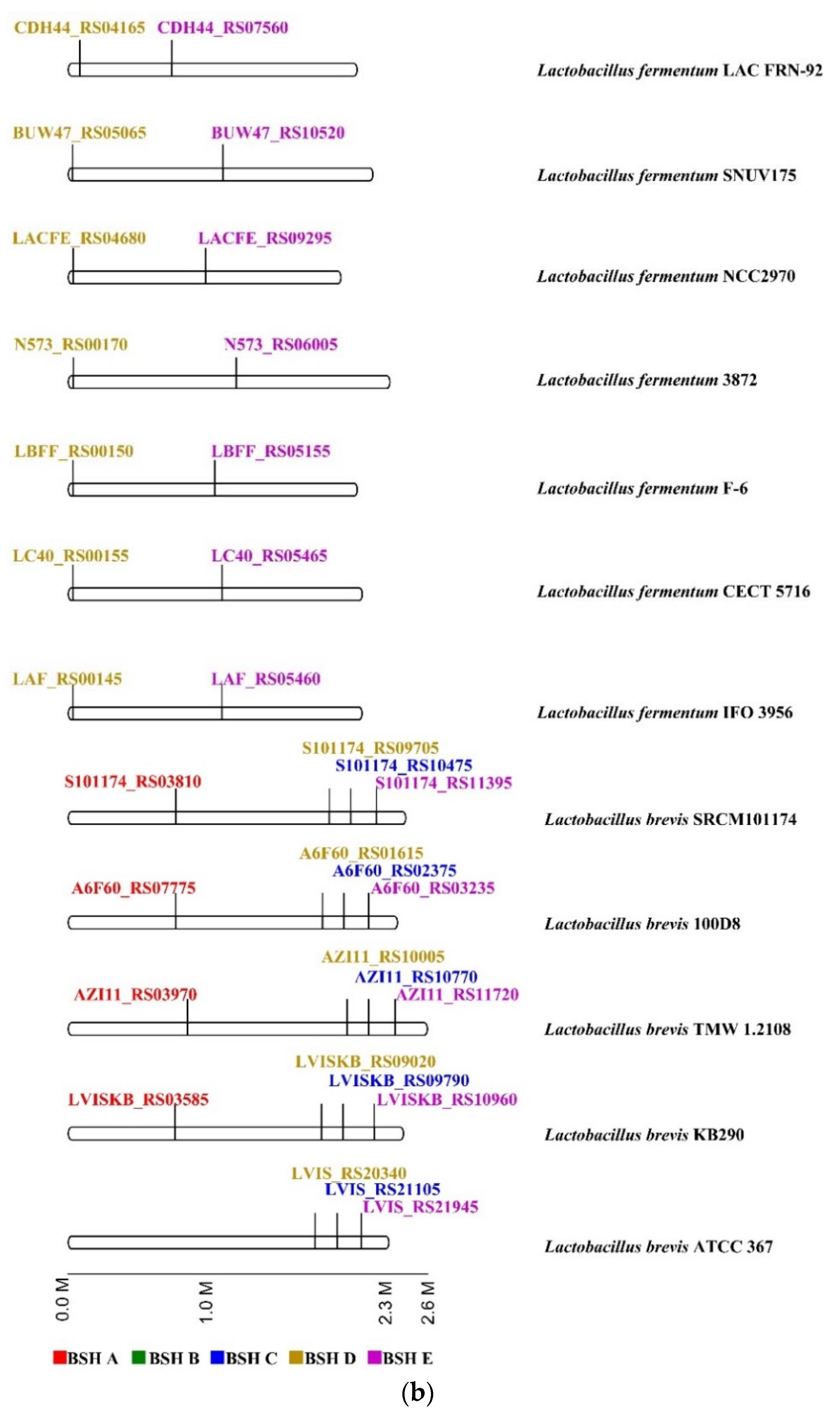
2.4. Genome Survey of bsh Genes in Lactobacillaceae Species
3. Discussion
4. Materials and Methods
4.1. Establishment of BSH–SDL
4.2. Identification of BSH in Lactobacillaceae
4.3. Genomic Location of bsh
4.4. Sequence Alignment and Phylogenetic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CVD | cardiovascular disease |
| TC | total cholesterol |
| LDL-C | low-density lipoprotein cholesterol |
| BAs | bile acids |
| CA | cholic acid |
| CDCA | chenodeoxycholic acid |
| TCA | taurocholic acid |
| TCDCA | taurochenodeoxycholic acid |
| GCA | glycocholic acid |
| GCDCA | glycochenodeoxycholic acid |
| BSH | bile salt hydrolase |
| BSH–SDL | bile salt hydrolase seed database of Lactobacillaceae |
| FXR | farnesoid X receptor |
| GPBAR1/TGR5 | G-protein-coupled bile acid receptor 1 |
References
- Lee, C.M.Y.; Barzi, F.; Woodward, M.; Batty, G.D.; Giles, G.G.; Wong, J.W.; Jamrozik, K.; Lam, T.H.; Ueshima, H.; Kim, H.C.; et al. Adult height and the risks of cardiovascular disease and major causes of death in the Asia-Pacific region: 21,000 deaths in 510,000 men and women. Int. J. Epidemiol. 2009, 38, 1060–1071. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Liu, X.M.; Zhang, Q.X.; Shen, Z.; Tian, F.W.; Zhang, H.; Sun, Z.H.; Zhang, H.P.; Chen, W. Influence of consumption of probiotics on the plasma lipid profile: A meta-analysis of randomised controlled trials. Nutr. Metab. Cardiovasc. Dis. 2011, 21, 844–850. [Google Scholar] [CrossRef] [PubMed]
- Anderson, K.E.; Kok, E.; Javitt, N.B. Bile acid synthesis in man: Metabolism of 7α-hydroxycholesterol-(14)C and 26-hydroxycholesterol-(3)H. J. Clin. Investig. 1972, 51, 112–117. [Google Scholar] [CrossRef] [PubMed]
- Jia, W.; Xie, G.; Jia, W. Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 111–128. [Google Scholar] [CrossRef] [PubMed]
- Allen, K.; Jaeschke, H.; Copple, B.L. Bile acids induce inflammatory genes in hepatocytes: A novel mechanism of inflammation during obstructive cholestasis. Am. J. Pathol. 2011, 178, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, H.; Bernstein, C.; Payne, C.M.; Dvorak, K. Bile acids as endogenous etiologic agents in gastrointestinal cancer. World J. Gastroenterol. WJG 2009, 15, 3329–3340. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, C.; Holubec, H.; Bhattacharyya, A.K.; Nguyen, H.; Payne, C.M.; Zaitlin, B.; Bernstein, H. Carcinogenicity of deoxycholate, a secondary bile acid. Arch. Toxicol. 2011, 85, 863–871. [Google Scholar] [CrossRef] [PubMed]
- Quante, M.; Bhagat, G.; Abrams, J.; Marache, F.; Good, P.; Lee, M.D.; Lee, Y.; Friedman, R.; Asfaha, S.; Dubeykovskaya, Z.; et al. Bile acid and inflammation activate gastric cardia stem cells in a mouse model of Barrett’s-like metaplasia. Cancer Cell 2012, 21, 36–51. [Google Scholar] [CrossRef] [PubMed]
- Xie, G.; Wang, X.; Huang, F.; Zhao, A.; Chen, W.; Yan, J.; Zhang, Y.; Lei, S.; Ge, K.; Zheng, X.; et al. Dysregulated hepatic bile acids collaboratively promote liver carcinogenesis. Int. J. Cancer 2016, 139, 1764–1775. [Google Scholar] [CrossRef] [PubMed]
- Molinaro, A.; Wahlström, A.; Marschall, H.-U. Role of bile acids in metabolic control. Trends Endocrinol. Metab. 2018, 29, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Wang, X.; Wang, J.; Wu, F.; Sui, Y.; Yang, L.; Wang, Z. Lactobacillus plantarum strains as potential probiotic cultures with cholesterol-lowering activity. J. Dairy Sci. 2013, 96, 2746–2753. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Nagpal, R.; Kumar, R.; Hemalatha, R.; Verma, V.; Kumar, A.; Chakraborty, C.; Singh, B.; Marotta, F.; Jain, S.; et al. Cholesterol-lowering probiotics as potential biotherapeutics for metabolic diseases. Exp. Diabetes Res. 2012, 2012, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ooi, L.-G.; Liong, M.-T. Cholesterol-lowering effects of probiotics and prebiotics: A review of in vivo and in vitro findings. Int. J. Mol. Sci. 2010, 11, 2499–2522. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, G.; Liu, X.-M.; Zhang, Q.-X.; Tian, F.-W.; Zhang, H.; Zhang, H.-P.; Chen, W. Research advances with regards to clinical outcome and potential mechanisms of the cholesterol-lowering effects of probiotics. Clin. Lipidol. 2012, 7, 501–507. [Google Scholar] [CrossRef]
- Chand, D.; Avinash, V.S.; Yadav, Y.; Pundle, A.V.; Suresh, C.G.; Ramasamy, S. Molecular features of bile salt hydrolases and relevance in human health. Biochim. Biophys. Acta 2017, 1861, 2981–2991. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.L.; Martoni, C.J.; Parent, M.; Prakash, S. Cholesterol-lowering efficacy of a microencapsulated bile salt hydrolase-active Lactobacillus reuteri NCIMB 30242 yoghurt formulation in hypercholesterolaemic adults. Br. J. Nutr. 2012, 107, 1505–1513. [Google Scholar] [CrossRef] [PubMed]
- Ejtahed, H.S.; Mohtadi-Nia, J.; Homayouni-Rad, A.; Niafar, M.; Asghari-Jafarabadi, M.; Mofid, V.; Akbarian-Moghari, A. Effect of probiotic yogurt containing Lactobacillus acidophilus and Bifidobacterium lactis on lipid profile in individuals with type 2 diabetes mellitus. J. Dairy Sci. 2011, 94, 3288–3294. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, M.C.; Lajo, T.; Carrión, J.M.; Cuñé, J. Cholesterol-lowering efficacy of Lactobacillus plantarum CECT 7527, 7528 and 7529 in hypercholesterolaemic adults. Br. J. Nutr. 2013, 109, 1866–1872. [Google Scholar] [CrossRef] [PubMed]
- Lambert, J.M.; Bongers, R.S.; de Vos, W.M.; Kleerebezem, M. Functional analysis of four bile salt hydrolase and penicillin acylase family members in Lactobacillus plantarum WCFS1. Appl. Environ. Microbiol. 2008, 74, 4719–4726. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Sun, K.; Wu, Z.; Yao, J.; Guo, B. All 4 bile salt hydrolase proteins are responsible for the hydrolysis activity in Lactobacillus plantarum ST-III. J. Food Sci. 2011, 76, M622–M628. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.S.; Brannigan, J.A.; Prabhune, A.A.; Pundle, A.V.; Dodson, G.G.; Dodson, E.J.; Suresh, C.G. Structural and functional analysis of a conjugated bile salt hydrolase from Bifidobacterium longum reveals an evolutionary relationship with penicillin V acylase. J. Biol. Chem. 2006, 281, 32516–32525. [Google Scholar] [CrossRef] [PubMed]
- McAuliffe, O.; Cano, R.J.; Klaenhammer, T.R. Genetic analysis of two bile salt hydrolase activities in Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 2005, 71, 4925–4929. [Google Scholar] [CrossRef] [PubMed]
- Jayashree, S.; Pooja, S.; Pushpanathan, M.; Rajendhran, J.; Gunasekaran, P. Identification and characterization of bile salt hydrolase genes from the genome of Lactobacillus fermentum MTCC 8711. Appl. Biochem. Biotechnol. 2014, 174, 855–866. [Google Scholar] [CrossRef] [PubMed]
- Asghar, S.; Arif, M.; Nawaz, M.; Muhammad, K.; Ali, M.A.; Ahmad, M.D.; Iqbal, S.; Anjum, A.A.; Khan, M.; Nazir, J. Selection, characterisation and evaluation of potential probiotic Lactobacillus spp. isolated from poultry droppings. Benef. Microbes 2016, 7, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Lodola, A.; Branduardi, D.; De Vivo, M.; Capoferri, L.; Mor, M.; Piomelli, D.; Cavalli, A. A Catalytic mechanism for cysteine N-Terminal nucleophile hydrolases, as revealed by free energy simulations. PLoS ONE 2012, 7, e32397. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Gu, Q. Complete genome sequence of Lactobacillus plantarum LZ95, a potential probiotic strain producing bacteriocins and B-group vitamin riboflavin. J. Biotechnol. 2016, 229, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Xiong, T.; Song, S.; Huang, X.; Feng, C.; Liu, G.; Huang, J.; Xie, M. Screening and identification of functional Lactobacillus specific for vegetable fermentation. J. Food Sci. 2012, 78, M84–M89. [Google Scholar] [CrossRef] [PubMed]
- Deraz, S.F.; Karlsson, E.N.; Khalil, A.A.; Mattiasson, B. Mode of action of acidocin D20079, a bacteriocin produced by the potential probiotic strain, Lactobacillus acidophilus DSM 20079. J. Ind. Microbiol. Biotechnol. 2007, 34, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Chae, J.P.; Valeriano, V.D.; Kim, G.-B.; Kang, D.-K. Molecular cloning, characterization and comparison of bile salt hydrolases from Lactobacillus johnsonii PF01. J. Appl. Microbiol. 2013, 114, 121–133. [Google Scholar] [CrossRef] [PubMed]
- Allain, T.; Chaouch, S.; Thomas, M.; Vallée, I.; Buret, A.G.; Langella, P.; Grellier, P.; Polack, B.; Bermúdez-Humarán, L.G.; Florent, I. Bile-salt-hydrolases from the probiotic strain Lactobacillus johnsonii La1 mediate anti-giardial activity in vitro and in vivo. Front. Microbiol. 2017, 8, 2707. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Wang, C.-L.; Sun, Y.; Li, A.-L.; Liu, F.; Meng, X.-C. Microencapsulation of Lactobacillus rhamnosus GG by transglutaminase cross-linked soy protein isolate to improve survival in simulated gastrointestinal conditions and yoghurt. J. Food Sci. 2016, 81, M1726–M1734. [Google Scholar] [CrossRef] [PubMed]
- Lecomte, V.; Kaakoush, N.O.; Maloney, C.A.; Raipuria, M.; Huinao, K.D.; Mitchell, H.M.; Morris, M.J. Changes in gut microbiota in rats fed a high fat diet correlate with obesity-associated metabolic parameters. PLoS ONE 2015, 10, e0126931. [Google Scholar] [CrossRef] [PubMed]
- Bi, J.; Fang, F.; Qiu, Y.; Yang, Q.; Chen, J. Substrate specificities of bile salt hydrolase 1 and its mutants from Lactobacillus salivarius. Sheng Wu Gong Cheng Xue Bao Chin. J. Biotechnol. 2014, 30, 445–454. [Google Scholar]
- Chen, C.; Xia, R.; Chen, H.; He, Y. TBtools, a toolkit for biologists integrating various HTS-data handling tools with a user-friendly interface. bioRxiv 2018. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Beitz, E. TeXshade: Shading and labeling of multiple sequence alignments using LaTeX2e. Bioinformatics 2000, 16, 135–139. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. ProtTest 3: Fast selection of best-fit models of protein evolution. Bioinformatics 2011, 27, 1164–1165. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available. |
| Species | BSH Subtypes | Tolerable Conditions | Substrate Specificities | Strains | |||||
|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | Bile Salt (%) | pH | |||
| Lactobacillus plantarum | 1 | 2 | 1 | 1.0 | 2.0 | GCA | ST-III [20] | ||
| 1 | 2 | 1 | - | - | GCA | WCFS1 [19] | |||
| 1 | 2 | 1 | 0.3 | 2.0 | - | LZ95 [26] | |||
| 1 | 1 | 1 | 1.0 | 2.0 | - | NCU116 [27] | |||
| Lactobacillus acidophilus | 2 | 0.2 | - | TCA, GCA, TCDCA | NCFM [22] | ||||
| 2 | 0.3 | 2.0 | - | DSM 20079 [28] | |||||
| Lactobacillus johnsonii | 2 | 0.5 | - | - | PF01 [29] | ||||
| 2 | 0.5 | 2.0 | GCDCA, TCDCA | La1 (also known as NCC533) [30] | |||||
| Lactobacillus rhamnosus | 1 | 2.0 | 2.5 | - | GG (ATCC 53103) [31] | ||||
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, L.; Yi, Y.; Lv, Y.; Qian, J.; Lei, X.; Zhang, G. A Comprehensive Genome Survey Provides Novel Insights into Bile Salt Hydrolase (BSH) in Lactobacillaceae. Molecules 2018, 23, 1157. https://doi.org/10.3390/molecules23051157
Liang L, Yi Y, Lv Y, Qian J, Lei X, Zhang G. A Comprehensive Genome Survey Provides Novel Insights into Bile Salt Hydrolase (BSH) in Lactobacillaceae. Molecules. 2018; 23(5):1157. https://doi.org/10.3390/molecules23051157
Chicago/Turabian StyleLiang, Lifeng, Yunhai Yi, Yunyun Lv, Junwei Qian, Xuejing Lei, and Gengyun Zhang. 2018. "A Comprehensive Genome Survey Provides Novel Insights into Bile Salt Hydrolase (BSH) in Lactobacillaceae" Molecules 23, no. 5: 1157. https://doi.org/10.3390/molecules23051157





