Identifying Cancer Specific Driver Modules Using a Network-Based Method
Abstract
:1. Introduction
2. Results
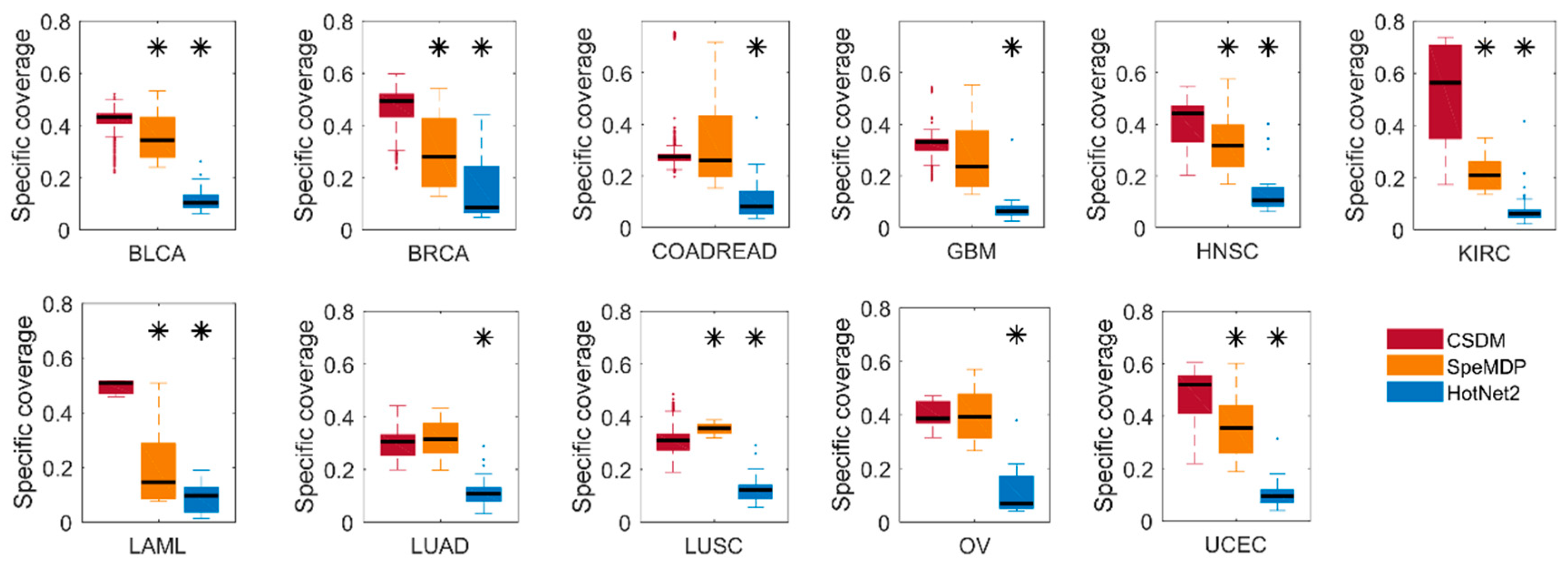
2.1. Comparison Study
2.1.1. Comparison of Specific Coverage
2.1.2. Comparison on Pathway Enrichment
2.2. Overlaps between Different Cancer Types
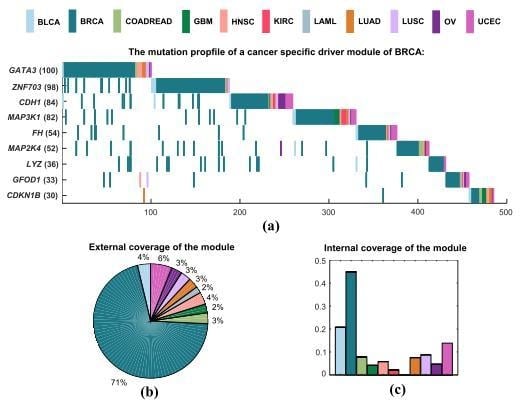
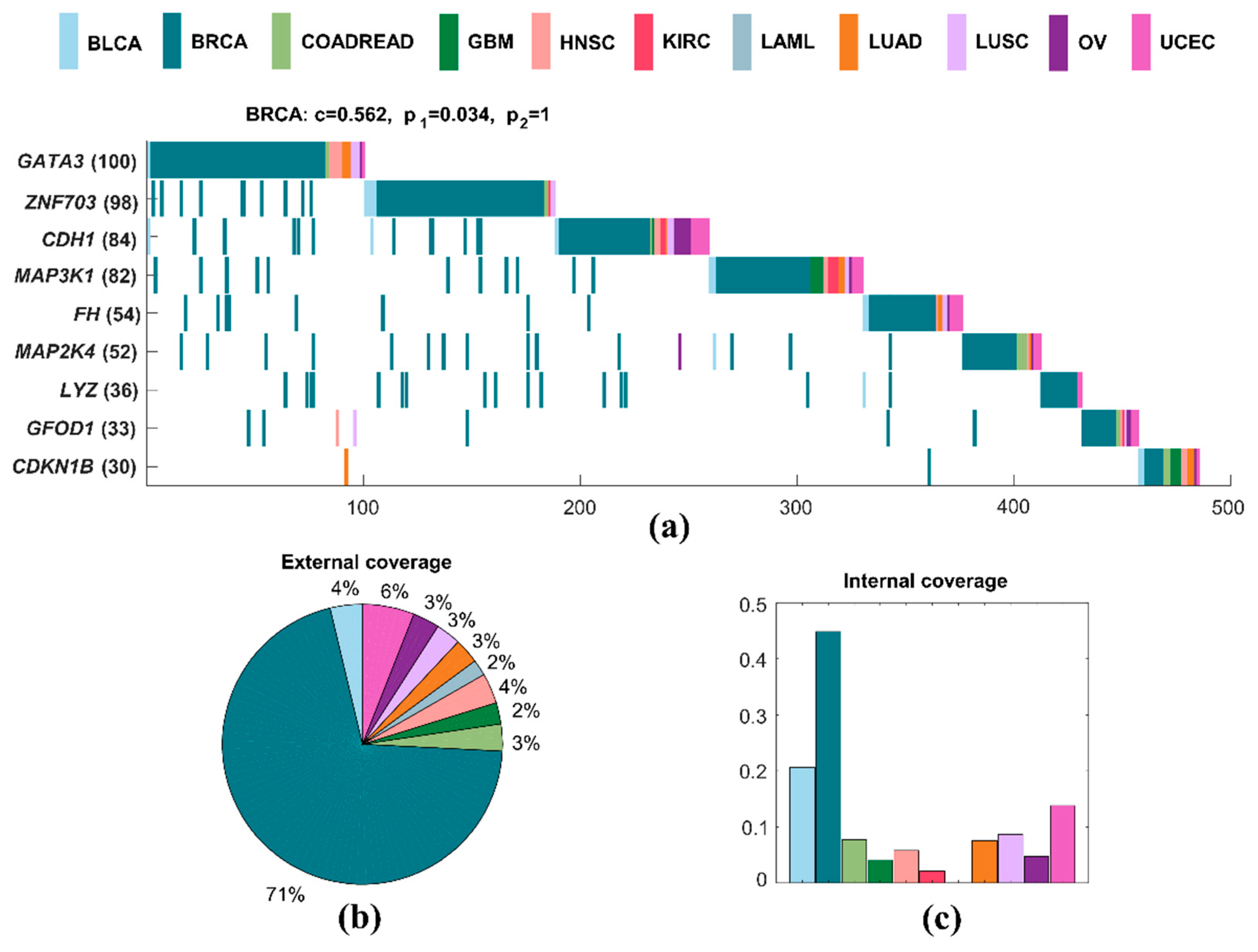
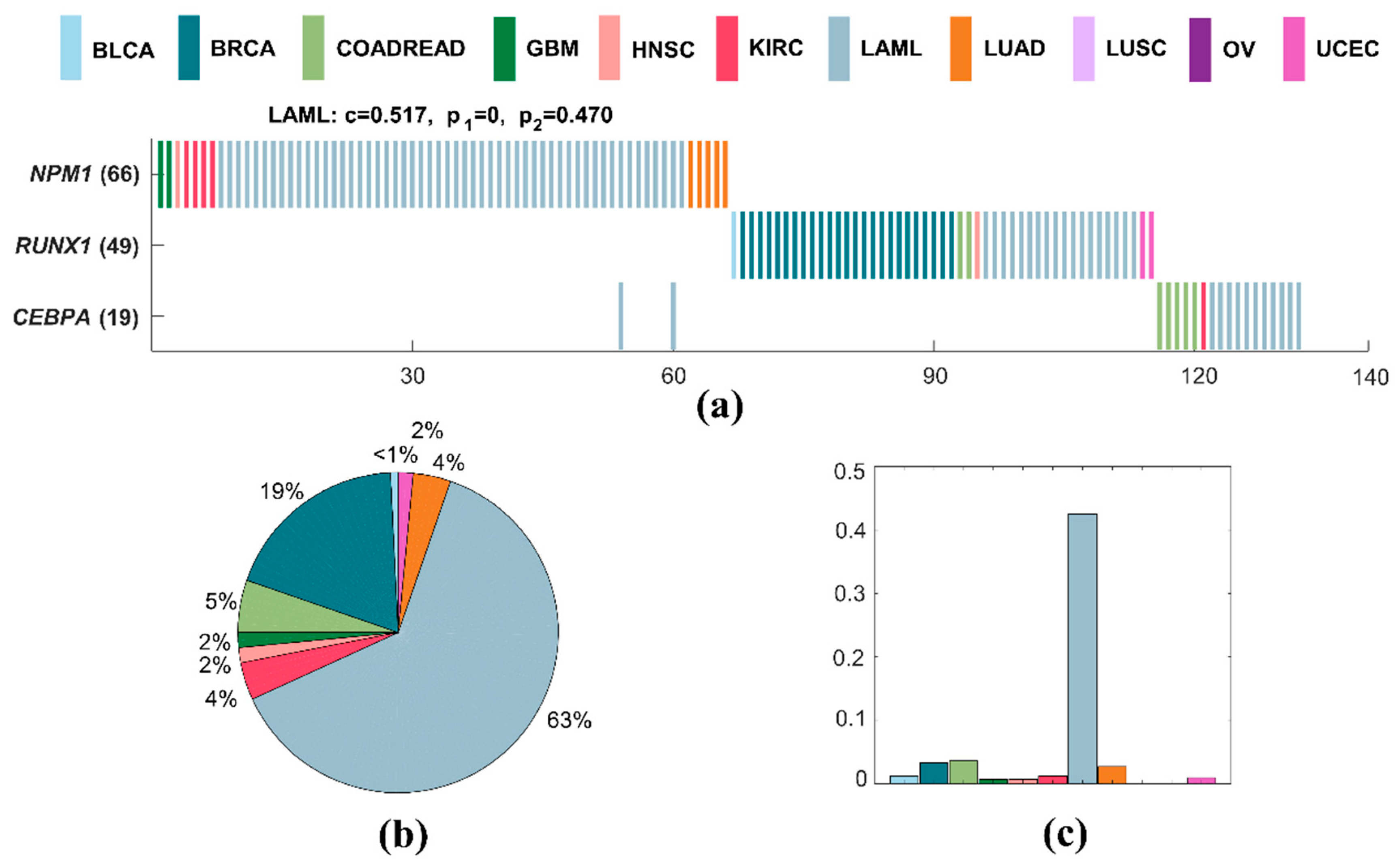
2.3. Specific Driver Modules in BRCA, BLCA, and LAML
2.3.1. Specific Driver Modules in BRCA
2.3.2. Specific Driver Modules in BLCA
2.3.3. Specific Driver Modules in LAML
3. Discussion
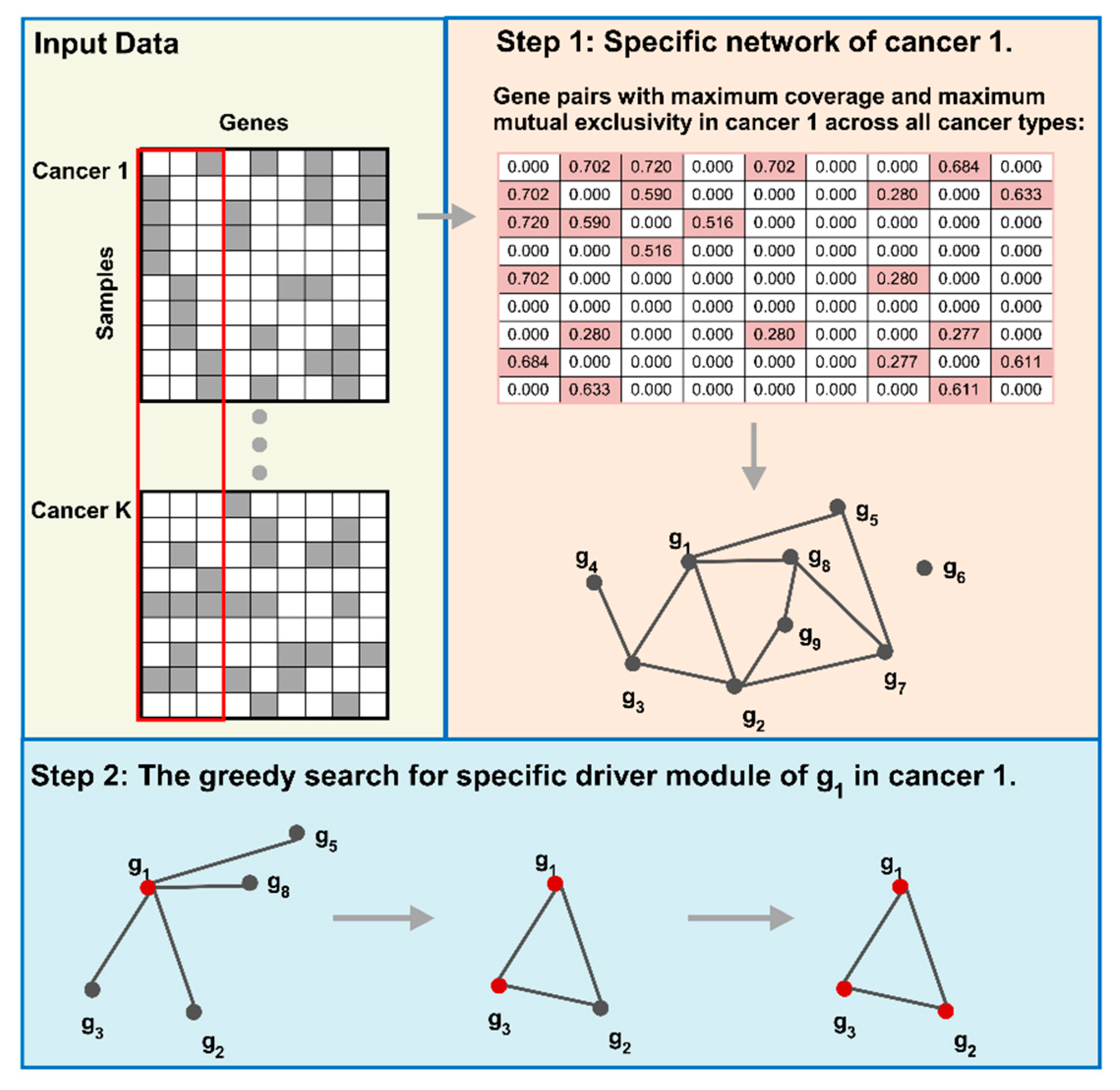
4. Datasets and Methods
4.1. Datasets
4.2. Methods
4.2.1. Cancer Specific Network Construction
4.2.2. Cancer Specific Driver Module Detection
| Algorithm 1 Cancer specific driver modules detection |
| Input: : mutation matrix of the th caner type; : mutation matrix for all cancer types; : specific network for cancer ; : a gene in network ; Output: : specific driver module of gene . Step 1: Initialize . Step 2: Compute objective function . (a) Compute the according to Equation (9). is the objective function. Step 3: Update and . (a) Compute the . is a set of neighbours of in , and is the number of neighbours. (b) Compute the according to Equation (9). is the external coverage of D and its neighbour gi. (c) Select the gene with and . (d) If exists, update and . Then go to Step 2. If does not exist, return . |
4.2.3. Evaluation Measures
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Vogelstein, B.; Papadopoulos, N.; Velculescu, V.E.; Zhou, S.; Diaz, L.A., Jr.; Kinzler, K.W. Cancer genome landscapes. Science 2013, 339, 1546–1558. [Google Scholar] [CrossRef] [PubMed]
- Dees, N.D.; Zhang, Q.; Kandoth, C.; Wendl, M.C.; Schierding, W.; Koboldt, D.C.; Mooney, T.B.; Callaway, M.B.; Dooling, D.; Mardis, E.R.; et al. Music: Identifying mutational significance in cancer genomes. Genome Res. 2012, 22, 1589–1598. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, M.S.; Stojanov, P.; Polak, P.; Kryukov, G.V.; Cibulskis, K.; Sivachenko, A.; Carter, S.L.; Stewart, C.; Mermel, C.H.; Roberts, S.A. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013, 499, 214–218. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, M.S.; Stojanov, P.; Mermel, C.H.; Robinson, J.T.; Garraway, L.A.; Golub, T.R.; Meyerson, M.; Gabriel, S.B.; Lander, E.S.; Getz, G. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 2014, 505, 495–501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reimand, J.; Bader, G.D. Systematic analysis of somatic mutations in phosphorylation signaling predicts novel cancer drivers. Mol. Syst. Biol. 2013, 9, 637. [Google Scholar] [CrossRef] [PubMed]
- Tamborero, D.; Gonzalez-Perez, A.; Lopez-Bigas, N. Oncodriveclust: Exploiting the positional clustering of somatic mutations to identify cancer genes. Bioinformatics 2013, 29, 2238–2244. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Perez, A.; Lopez-Bigas, N. Functional impact bias reveals cancer drivers. Nucleic Acids Res. 2012, 40, e169. [Google Scholar] [CrossRef] [PubMed]
- Mularoni, L.; Sabarinathan, R.; Deu-Pons, J.; Gonzalez-Perez, A.; López-Bigas, N. Oncodrivefml: A general framework to identify coding and non-coding regions with cancer driver mutations. Genome Biol. 2016, 17, 128. [Google Scholar] [CrossRef] [PubMed]
- Davoli, T.; Xu, A.W.; Mengwasser, K.E.; Sack, L.M.; Yoon, J.C.; Park, P.J.; Elledge, S.J. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell 2013, 155, 948–962. [Google Scholar] [CrossRef] [PubMed]
- Tokheim, C.J.; Papadopoulos, N.; Kinzler, K.W.; Vogelstein, B.; Karchin, R. Evaluating the evaluation of cancer driver genes. Proc. Natl. Acad. Sci. USA 2016, 113, 14330–14335. [Google Scholar] [CrossRef] [PubMed]
- Bailey, M.H.; Tokheim, C.; Porta-Pardo, E.; Sengupta, S.; Bertrand, D.; Weerasinghe, A.; Colaprico, A.; Wendl, M.C.; Kim, J.; Reardon, B.; et al. Comprehensive characterization of cancer driver genes and mutations. Cell 2018, 173, 371–385.e318. [Google Scholar] [CrossRef] [PubMed]
- Cho, A.; Shim, J.E.; Kim, E.; Supek, F.; Lehner, B.; Lee, I. Muffinn: Cancer gene discovery via network analysis of somatic mutation data. Genome Biol. 2016, 17, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Shi, K.; Gao, L.; Wang, B. Discovering potential cancer driver genes by an integrated network-based approach. Mol. BioSyst. 2016, 12, 2921–2931. [Google Scholar] [CrossRef] [PubMed]
- Xi, J.; Li, A.; Wang, M. A novel network regularized matrix decomposition method to detect mutated cancer genes in tumour samples with inter-patient heterogeneity. Sci. Rep. 2017, 7, 2855. [Google Scholar] [CrossRef] [PubMed]
- Xi, J.; Wang, M.; Li, A. Discovering potential driver genes through an integrated model of somatic mutation profiles and gene functional information. Mol. BioSyst. 2017, 13, 2135–2144. [Google Scholar] [CrossRef] [PubMed]
- Xi, J.; Wang, M.; Li, A. Dgpathinter: A novel model for identifying driver genes via knowledge-driven matrix factorization with prior knowledge from interactome and pathways. PeerJ Comput. Sci. 2017, 3, e133. [Google Scholar] [CrossRef]
- Wang, Y.-X.; Liu, J.-X.; Gao, Y.-L.; Zheng, C.-H.; Shang, J.-L. Differentially expressed genes selection via laplacian regularized low-rank representation method. Comput. Biol. Chem. 2016, 65, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.Y.; Liu, J.X.; Zheng, C.H.; Shang, J.; Feng, C.M.; Wang, Y.X. Robust graph regularized sparse orthogonal nonnegative matrix factorization for identifying differentially expressed genes. In Proceedings of the 2017 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Kansas City, MO, USA, 13–16 November 2017; pp. 1900–1905. [Google Scholar]
- Ding, L.; Getz, G.; Wheeler, D.A.; Mardis, E.R.; McLellan, M.D.; Cibulskis, K.; Sougnez, C.; Greulich, H.; Muzny, D.M.; Morgan, M.B. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 2008, 455, 1069–1075. [Google Scholar] [CrossRef] [PubMed]
- Network, C.G.A.R. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008, 455, 1061–1068. [Google Scholar]
- Yeang, C.-H.; McCormick, F.; Levine, A. Combinatorial patterns of somatic gene mutations in cancer. FASEB J. 2008, 22, 2605–2622. [Google Scholar] [CrossRef] [PubMed]
- Vogelstein, B.; Kinzler, K.W. Cancer genes and the pathways they control. Nat. Med. 2004, 10, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Dimitrakopoulos, C.M.; Beerenwinkel, N. Computational approaches for the identification of cancer genes and pathways. Wiley Interdiscip. Rev. Syst. Biol. Med. 2017, 9, e1364. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, S. The discovery of mutated driver pathways in cancer: Models and algorithms. IEEE/ACM Trans. Comput. Biol. Bioinform. 2017, 1. [Google Scholar] [CrossRef] [PubMed]
- Vandin, F.; Upfal, E.; Raphael, B.J. De novo discovery of mutated driver pathways in cancer. Genome Res. 2012, 22, 375–385. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhang, S.; Wu, L.-Y.; Zhang, X.-S. Efficient methods for identifying mutated driver pathways in cancer. Bioinformatics 2012, 28, 2940–2947. [Google Scholar] [CrossRef] [PubMed]
- Leiserson, M.D.M.; Blokh, D.; Sharan, R.; Raphael, B.J. Simultaneous identification of multiple driver pathways in cancer. PLoS Comput. Biol. 2013, 9, e1003054. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, S.; Wang, Y.; Zhang, X.S. Identification of mutated core cancer modules by integrating somatic mutation, copy number variation, and gene expression data. BMC Syst. Biol. 2013, 7 (Suppl. S2), S4. [Google Scholar] [CrossRef] [PubMed]
- Constantinescu, S.; Szczurek, E.; Mohammadi, P.; Rahnenführer, J.; Beerenwinkel, N. Timex: A waiting time model for mutually exclusive cancer alterations. Bioinformatics 2015, 32, 968–975. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-A.; Madan, S.; Przytycka, T.M. Wesme: Uncovering mutual exclusivity of cancer drivers and beyond. Bioinformatics 2016, 33, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Gao, L.; Ma, X.; Yang, X. Detection of driver pathways using mutated gene network in cancer. Mol. BioSyst. 2016, 12, 2135–2141. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, S. Discovery of cancer common and specific driver gene sets. Nucleic Acids Res. 2017, 45, e86. [Google Scholar] [CrossRef] [PubMed]
- Vandin, F.; Upfal, E.; Raphael, B.J. Algorithms for detecting significantly mutated pathways in cancer. J. Comput. Biol. 2011, 18, 507–522. [Google Scholar] [CrossRef] [PubMed]
- Ciriello, G.; Cerami, E.; Sander, C.; Schultz, N. Mutual exclusivity analysis identifies oncogenic network modules. Genome Res. 2012, 22, 398–406. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-A.; Cho, D.-Y.; Dao, P.; Przytycka, T.M. Memcover: Integrated analysis of mutual exclusivity and functional network reveals dysregulated pathways across multiple cancer types. Bioinformatics 2015, 31, i284–i292. [Google Scholar] [CrossRef] [PubMed]
- Leiserson, M.D.M.; Vandin, F.; Wu, H.-T.; Dobson, J.R.; Eldridge, J.V.; Thomas, J.L.; Papoutsaki, A.; Kim, Y.; Niu, B.; McLellan, M.; et al. Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nat. Genet. 2015, 47, 106–114. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Vega, F.; Mina, M.; Armenia, J.; Chatila, W.K.; Luna, A.; La, K.C.; Dimitriadoy, S.; Liu, D.L.; Kantheti, H.S.; Saghafinia, S.; et al. Oncogenic signaling pathways in the cancer genome atlas. Cell 2018, 173, 321–337. [Google Scholar] [CrossRef] [PubMed]
- Ge, Z.; Leighton, J.S.; Wang, Y.; Peng, X.; Chen, Z.; Chen, H.; Sun, Y.; Yao, F.; Li, J.; Zhang, H.; et al. Integrated genomic analysis of the ubiquitin pathway across cancer types. Cell Rep. 2018, 23, 213–226.e213. [Google Scholar] [CrossRef] [PubMed]
- Schaub, F.X.; Dhankani, V.; Berger, A.C.; Trivedi, M.; Richardson, A.B.; Shaw, R.; Zhao, W.; Zhang, X.; Ventura, A.; Liu, Y.; et al. Pan-cancer alterations of the myc oncogene and its proximal network across the cancer genome atlas. Cell Syst. 2018, 6, 282–300.e282. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Tang, W.; Wang, P.; Guo, X.; Gao, L. Extracting stage-specific and dynamic modules through analyzing multiple networks associated with cancer progression. IEEE/ACM Trans. Comput. Biol. Bioinform. 2016, 647–658. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Shao, X.; Gao, L.; Zhang, S. Comparative DNA methylation analysis to decipher common and cell type-specific patterns among multiple cell types. Brief. Funct. Genom. 2016, 15, 399–407. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Sun, P.; Qin, G. Identifying condition-specific modules by clustering multiple networks. IEEE/ACM Trans. Comput. Biol. Bioinform. 2017, 1. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Gao, L.; Zhang, S. Comparative pan-cancer DNA methylation analysis reveals cancer common and specific patterns. Brief. Bioinform. 2017, 18, 761–773. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Sato, Y.; Furumichi, M.; Tanabe, M. Kegg for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012, 40, D109–D114. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The molecular signatures database hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, C.F.; Anthony, K.; Krupa, S.; Buchoff, J.; Day, M.; Hannay, T.; Buetow, K.H. Pid: The pathway interaction database. Nucleic Acids Res. 2009, 37, D674–D679. [Google Scholar] [CrossRef] [PubMed]
- Fabregat, A.; Sidiropoulos, K.; Garapati, P.; Gillespie, M.; Hausmann, K.; Haw, R.; Jassal, B.; Jupe, S.; Korninger, F.; McKay, S.; et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2016, 44, D481–D487. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. Kegg: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Creighton, C.J.; Massarweh, S.; Huang, S.; Tsimelzon, A.; Hilsenbeck, S.G.; Osborne, C.K.; Shou, J.; Malorni, L.; Schiff, R. Development of resistance to targeted therapies transforms the clinically associated molecular profile subtype of breast tumor xenografts. Cancer Res. 2008, 68, 7493. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.-R.; Chen, Y.-H.; Chen, W.; Kong, W.; Huang, J.-W.; Zhang, J.; Xue, W.; Liu, D.-M.; Huang, Y.-R. GFOD1 and peejar are promising markers for clear-cell renal cell carcinoma disease progression. Oncotarget 2016, 7, 38004–38009. [Google Scholar] [CrossRef] [PubMed]
- Alaimo, S.; Giugno, R.; Acunzo, M.; Veneziano, D.; Ferro, A.; Pulvirenti, A. Post-transcriptional knowledge in pathway analysis increases the accuracy of phenotypes classification. Oncotarget 2016, 7, 54572–54582. [Google Scholar] [CrossRef] [PubMed]
- Draghici, S.; Khatri, P.; Tarca, A.L.; Amin, K.; Done, A.; Voichita, C.; Georgescu, C.; Romero, R. A systems biology approach for pathway level analysis. Genome Res. 2007, 17, 1537–1545. [Google Scholar] [CrossRef] [PubMed]
- Tarca, A.L.; Draghici, S.; Khatri, P.; Hassan, S.S.; Mittal, P.; Kim, J.-S.; Kim, C.J.; Kusanovic, J.P.; Romero, R. A novel signaling pathway impact analysis. Bioinformatics 2009, 25, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Alaimo, S.; Marceca, P.G.; Ferro, A.; Pulvirenti, A. Detecting disease specific pathway substructures through an integrated systems biology approach. Non-Coding RNA 2017, 3, 20. [Google Scholar] [CrossRef] [PubMed]
- The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014, 507, 315–322. [Google Scholar]
- Udayakumar, T.S.; Belakavadi, M.; Choi, K.-H.; Pandey, P.K.; Fondell, J.D. Regulation of aurora-a kinase gene expression via GABP recruitment of TRAP220/MED1. J. Biol. Chem. 2006, 281, 14691–14699. [Google Scholar] [CrossRef] [PubMed]
- Winter, S.C.; Buffa, F.M.; Silva, P.; Miller, C.; Valentine, H.R.; Turley, H.; Shah, K.A.; Cox, G.J.; Corbridge, R.J.; Homer, J.J.; et al. Relation of a hypoxia metagene derived from head and neck cancer to prognosis of multiple cancers. Cancer Res. 2007, 67, 3441–3449. [Google Scholar] [CrossRef] [PubMed]
- Döhner, H.; Weisdorf, D.J.; Bloomfield, C.D. Acute myeloid leukemia. N. Engl. J. Med. 2015, 373, 1136–1152. [Google Scholar] [CrossRef] [PubMed]
- Gill, H.; Leung, A.Y.H.; Kwong, Y.-L. Molecularly targeted therapy in acute myeloid leukemia. Future Oncol. 2016, 12, 827–838. [Google Scholar] [CrossRef] [PubMed]
- Chiu, Y.C.; Tsai, M.H.; Chou, W.C.; Liu, Y.C.; Kuo, Y.Y.; Hou, H.A.; Lu, T.P.; Lai, L.C.; Chen, Y.; Tien, H.F.; et al. Prognostic significance of NPM1 mutation-modulated microrna−mrna regulation in acute myeloid leukemia. Leukemia 2015, 30, 274–284. [Google Scholar] [CrossRef] [PubMed]
- Alpermann, T.; Schnittger, S.; Eder, C.; Dicker, F.; Meggendorfer, M.; Kern, W.; Schmid, C.; Aul, C.; Staib, P.; Wendtner, C.-M.; et al. Molecular subtypes of NPM1 mutations have different clinical profiles, specific patterns of accompanying molecular mutations and varying outcomes in intermediate risk acute myeloid leukemia. Haematologica 2016, 101, e55. [Google Scholar] [CrossRef] [PubMed]
- Gaidzik, V.I.; Bullinger, L.; Schlenk, R.F.; Zimmermann, A.S.; Röck, J.; Paschka, P.; Corbacioglu, A.; Krauter, J.; Schlegelberger, B.; Ganser, A.; et al. RUNX1 mutations in acute myeloid leukemia: Results from a comprehensive genetic and clinical analysis from the aml study group. J. Clin. Oncol. 2011, 29, 1364–1372. [Google Scholar] [CrossRef] [PubMed]
- Taskesen, E.; Bullinger, L.; Corbacioglu, A.; Sanders, M.A.; Erpelinck, C.A.J.; Wouters, B.J.; van der Poel-van de Luytgaarde, S.C.; Damm, F.; Krauter, J.; Ganser, A.; et al. Prognostic impact, concurrent genetic mutations, and gene expression features of aml with cebpa mutations in a cohort of 1182 cytogenetically normal aml patients: Further evidence for cebpa double mutant aml as a distinctive disease entity. Blood 2011, 117, 2469–2475. [Google Scholar] [CrossRef] [PubMed]
- Greshock, J.; Nathanson, K.; Martin, A.-M.; Zhang, L.; Coukos, G.; Weber, B.L.; Zaks, T.Z. Cancer cell lines as genetic models of their parent histology: Analyses based on array comparative genomic hybridization. Cancer Res. 2007, 67, 3594–3600. [Google Scholar] [CrossRef] [PubMed]
- Numata, A.; Shimoda, K.; Kamezaki, K.; Haro, T.; Kakumitsu, H.; Shide, K.; Kato, K.; Miyamoto, T.; Yamashita, Y.; Oshima, Y.; et al. Signal transducers and activators of transcription 3 augments the transcriptional activity of ccaat/enhancer-binding protein α in granulocyte colony-stimulating factor signaling pathway. J. Biol. Chem. 2005, 280, 12621–12629. [Google Scholar] [CrossRef] [PubMed]
- Bowers, P.M.; Cokus, S.J.; Eisenberg, D.; Yeates, T.O. Use of logic relationships to decipher protein network organization. Science 2004, 306, 2246–2249. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar]
Sample Availability: Samples of the compounds are available from the authors. |


| Pathway | Impact Factor | p-Value |
|---|---|---|
| Pathways in cancer | 11.282 | 2.134 × 10−5 |
| Epithelial cell signaling in Helicobacter pylori infection | 4.468 | 8.044 × 10−4 |
| Hepatitis B | 13.121 | 8.987 × 10−4 |
| Epstein–Barr virus infection | 7.806 | 1.101 × 10−3 |
| HTLV-I infection | 6.945 | 1.259 × 10−3 |
| GnRH signaling pathway | 9.044 | 2.054 × 10−3 |
| ErbB signaling pathway | 9.222 | 2.631 × 10−3 |
| Small cell lung cancer | 4.925 | 2.803 × 10−3 |
| Neurotrophin signaling pathway | 4.139 | 7.703 × 10−3 |
| Pathway | Impact Factor | p-Value |
|---|---|---|
| Pathways in cancer | 16.834 | 4.09 × 10−8 |
| Melanoma | 14.011 | 5.52 × 10−4 |
| Glioma | 14.099 | 6.58 × 10−4 |
| Chronic myeloid leukemia | 13.983 | 6.62 × 10−4 |
| PI3K–Akt signaling pathway | 11.242 | 8.61 × 10−4 |
| HTLV-I infection | 12.696 | 1.36 × 10−3 |
| Proteoglycans in cancer | 10.732 | 3.48 × 10−3 |
| Hepatitis B | 10.318 | 4.63 × 10−3 |
| p53 signaling pathway | 11.093 | 7.50 × 10−3 |
| PPAR signaling pathway | 5.895 | 9.08 × 10−3 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, F.; Gao, L.; Wang, P.; Hu, Y. Identifying Cancer Specific Driver Modules Using a Network-Based Method. Molecules 2018, 23, 1114. https://doi.org/10.3390/molecules23051114
Li F, Gao L, Wang P, Hu Y. Identifying Cancer Specific Driver Modules Using a Network-Based Method. Molecules. 2018; 23(5):1114. https://doi.org/10.3390/molecules23051114
Chicago/Turabian StyleLi, Feng, Lin Gao, Peizhuo Wang, and Yuxuan Hu. 2018. "Identifying Cancer Specific Driver Modules Using a Network-Based Method" Molecules 23, no. 5: 1114. https://doi.org/10.3390/molecules23051114