Synthesis and Biological Evaluation of Azamacrolide Comprising the Triazole Moiety as Quorum Sensing Inhibitors
Abstract
:1. Introduction
2. Results and Discussion
2.1. Chemistry
2.2. QS Activity.
2.2.1. QS Inhibitory Activity
2.2.2. Dose–Response Bioassay
2.2.3. Non-Competitive Inhibition Assay
2.2.4. QS Agonist
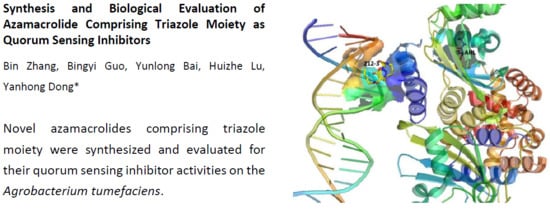
2.3. Docking
3. Experimental
3.1. QS Inhibitory Activity
3.2. The Bactericidal Activities of the Compounds
3.3. Noncompetition Assay
3.4. QS Agonist
3.5. Computational Chemistry—Docking
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Forschner-Dancause, S.; Poulin, E.; Meschwitz, S. Quorum Sensing Inhibition and Structure–Activity Relationships of β-Keto Esters. Molecules 2016, 21, 971. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, A.; Månsson, M.; Bojer, M.S.; Gram, L.; Larsen, T.O.; Novick, R.P.; Frees, D.; Frøkiær, H.; Ingmer, H. Solonamide B inhibits quorum sensing and reduces Staphylococcus aureus mediated killing of human neutrophils. PLoS ONE 2014, 9, e84992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vermote, A.; Brackman, G.; Risseeuw, M.D.; Vanhoutte, B.; Cos, P.; Van Hecke, K.; Breyne, K.; Meyer, E.; Coenye, T.; Van Calenbergh, S. Hamamelitannin Analogues that Modulate Quorum Sensing as Potentiators of Antibiotics against Staphylococcus aureus. Angew. Chem. Int. Ed. 2016, 55, 6551–6555. [Google Scholar] [CrossRef] [PubMed]
- Vermote, A.; Brackman, G.; Risseeuw, M.D.; Coenye, T.; Van Calenbergh, S. Novel hamamelitannin analogues for the treatment of biofilm related MRSA infections—A scaffold hopping approach. Eur. J. Med. Chem. 2017, 127, 757–770. [Google Scholar] [CrossRef] [PubMed]
- Suga, H.; Smith, K.M. Molecular mechanisms of bacterial quorum sensing as a new drug target. Curr. Opin. Chem. Biol. 2003, 7, 586–591. [Google Scholar] [CrossRef] [PubMed]
- Katz, L.; Ashley, G.W. Translation and Protein Synthesis: Macrolides. Chem. Rev. 2005, 105, 499–527. [Google Scholar] [CrossRef] [PubMed]
- Agouridas, C.; Denis, A.; Auger, J.M.; Benedetti, Y.; Bonnefoy, A.; Bretin, F.; Chantot, J.-F.; Dussarat, A.; Fromentin, C.; D’Ambrières, S.G.; et al. Synthesis and Antibacterial Activity of Ketolides (6-O-Methyl-3-oxoerythromycin Derivatives): A New Class of Antibacterials Highly Potent Against Macrolide-Resistant and -Susceptible Respiratory Pathogens. J. Med. Chem. 1998, 41, 4080–4100. [Google Scholar] [CrossRef] [PubMed]
- Swatton, J.E.; Davenport, P.W.; Maunders, E.A.; Griffin, J.L.; Lilley, K.S.; Welch, M. Impact of Azithromycin on the Quorum Sensing-Controlled Proteome of Pseudomonas aeruginosa. PLoS ONE 2016, 11, e0147698. [Google Scholar] [CrossRef] [PubMed]
- Tateda, K.; Comte, R.; Pechere, J.C.; Köhler, T.; Yamaguchi, K.; Van Delden, C. Azithromycin inhibits quorum sensing in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2001, 45, 1930–1933. [Google Scholar] [CrossRef] [PubMed]
- Saiman, L.; Chen, Y.; San Gabriel, P.; Knirsch, C. Synergistic Activities of Macrolide Antibiotics against Pseudomonas aeruginosa, Burkholderia cepacia, Stenotrophomonas maltophilia, and Alcaligenes xylosoxidans Isolated from Patients with Cystic Fibrosis. Antimicrob. Agents Chemother. 2002, 46, 1105–1107. [Google Scholar] [CrossRef] [PubMed]
- Bratu, S.; Quale, J.; Cebular, S.; Heddurshetti, R.; Landman, D. Multidrug-resistant Pseudomonas aeruginosa in Brooklyn, New York: Molecular epidemiology and in vitro activity of polymyxin B. Eur. J. Clin. Microbiol. Infect. Dis. 2005, 24, 196–201. [Google Scholar] [CrossRef] [PubMed]
- Senwar, K.R.; Sharma, P.; Reddy, T.S.; Jeengar, M.K.; Nayak, V.L.; Naidu, V.G.M.; Kamal, A.; Shankaraiah, N. Spirooxindole-derived morpholine-fused-1,2,3-triazoles: Design, synthesis, cytotoxicity and apoptosis inducing studies. Eur. J. Med. Chem. 2015, 102, 413–424. [Google Scholar] [CrossRef] [PubMed]
- Fu, O.; Pukin, A.V.; Quarles van Ufford, H.C.; Kemmink, J.; de Mol, N.J.; Pieters, R.J. Functionalization of a Rigid Divalent Ligand for LecA, a Bacterial Adhesion Lectin. ChemistryOpen 2015, 4, 463–470. [Google Scholar] [CrossRef] [PubMed]
- Fraley, M.E.; Steen, J.T.; Brnardic, E.J.; Arrington, K.L.; Spencer, K.L.; Hanney, B.A.; Kim, Y.; Hartman, G.D.; Stirdivant, S.M.; Drakas, B.A.; et al. 3-(Indol-2-yl)indazoles as Chek1 kinase inhibitors: Optimization of potency and selectivity via substitution at C6. Bioorg. Med. Chem. Lett. 2006, 16, 6049–6053. [Google Scholar] [CrossRef] [PubMed]
- Hansen, M.R.; Jakobsen, T.H.; Bang, C.G.; Cohrt, A.E.; Hansen, C.L.; Clausen, J.W.; Le Quement, S.T.; Tolker-Nielsen, T.; Givskov, M.; Nielsen, T.E. Triazole-containing N-acyl homoserine lactones targeting the quorum sensing system in Pseudomonas aeruginosa. Bioorg. Med. Chem. 2015, 23, 1638–1650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, D.W.; Zhang, Y.M.; Li, J.; Zhao, T.Q.; Gu, Q.; Lin, F. Ultrasonic-assisted synthesis of 1,4-disubstituted 1,2,3-triazoles via various terminal acetylenes and azide and their quorum sensing inhibition. Ultrason. Sonochem. 2017, 36, 343–353. [Google Scholar] [CrossRef] [PubMed]
- Viegas-Junior, C.; Danuello, A.; da Silva Bolzani, V.; Barreiro, E.J.; Fraga, C.A.M. Molecular hybridization: A useful tool in the design of new drug prototypes. Curr. Med. Chem. 2007, 14, 1829–1852. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Rahman, S.A.; El-Gohary, N.S.; El-Bendary, E.R.; El-Ashry, S.M.; Shaaban, M.I. Synthesis, antimicrobial, antiquorum-sensing, antitumor and cytotoxic activities of new series of cyclopenta(hepta)[b]thiophene and fused cyclohepta[b]thiophene analogs. Eur. J. Med. Chem. 2017, 140, 200–211. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.G.; Pappas, K.M.; Brace, J.L.; Miller, P.C.; Oulmassov, T.; Molyneaux, J.M.; Anderson, J.C.; Bashkin, J.K.; Winans, S.C.; Joachimiak, A. Structure of a bacterial quorum-sensingtranscription factor complexed with pheromone and DNA. Nature 2002, 417, 971–974. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Liang, X.; Yuan, H.; Qi, S.; Chen, F.; Wang, D. Potential green fungicide: 16-oxo-1-oxa-4-azoniacyclohexadecan-4-ium tetrafluoroborate. Green Chem. 2008, 10, 990–994. [Google Scholar] [CrossRef]
- King, E.O.; Ward, M.K.; Raney, D.E. Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 1954, 44, 301–307. [Google Scholar] [PubMed]
- Erdönmez, D.; Rad, A.Y.; Aksöz, N. Quorum sensing molecules production by nosocomial and soil isolates Acinetobacter baumannii. Arch. Microbiol. 2017, 199, 1325–1334. [Google Scholar] [CrossRef] [PubMed]
- Gui, M.; Wu, R.; Liu, L.; Wang, S.; Zhang, L.; Li, P. Effects of quorum quenching by AHL lactonase on AHLs, protease, motility and proteome patterns in Aeromonas veronii LP-11. Int. J. Food Microbiol. 2017, 252, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Liang, L.; Zhu, Y.; Qiu, S.; Wang, T.; Zhang, L. Ligand and Structure-Based Approaches for the Identification of Peptide Deformylase Inhibitors as Antibacterial Drugs. Int. J. Mol. Sci. 2016, 17, 1141. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Gao, P.; Liu, Y.; Ji, X.; Gan, M.; Guan, Y.; Hao, X.; Li, Z.; Xiao, C. A discovery of novel Mycobacterium tuberculosis pantothenate synthetase inhibitors based on the molecular mechanism of actinomycin D inhibition. Bioorg. Med. Chem. Lett. 2011, 21, 3943–3946. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Not Available. |









| Comp. | OD600 | Inhibition (%) | Comp. | OD600 | Inhibition (%) |
|---|---|---|---|---|---|
| Blank | 0.664 | --- | DMSO | 0.683 | −2.2 |
| Z12-2 | 0.73 | −9.9 | Z16-3 | 0.838 | −25.5 |
| Z12-9 | 0.826 | −22.2 | Z16-9 | 0.845 | −21.6 |
| Z13-1 | 0.811 | −17.8 | Z16-10 | 0.868 | −24.1 |
| Z13-3 | 0.85 | −22.9 | Z16-11 | 0.815 | −17.4 |
| Group | U | Inhibition (%) |
|---|---|---|
| signal | 0.47 | —— |
| signal + 100 mg/L | 0.46 | 1.05 |
| signal + 50 mg/L | 0.78 | −66.67 |
| 100 mg/L | 0.04 | —— |
| 50 mg/L | 0.03 | —— |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, B.; Guo, B.; Bai, Y.; Lu, H.; Dong, Y. Synthesis and Biological Evaluation of Azamacrolide Comprising the Triazole Moiety as Quorum Sensing Inhibitors. Molecules 2018, 23, 1086. https://doi.org/10.3390/molecules23051086
Zhang B, Guo B, Bai Y, Lu H, Dong Y. Synthesis and Biological Evaluation of Azamacrolide Comprising the Triazole Moiety as Quorum Sensing Inhibitors. Molecules. 2018; 23(5):1086. https://doi.org/10.3390/molecules23051086
Chicago/Turabian StyleZhang, Bin, Bingyi Guo, Yunlong Bai, Huizhe Lu, and Yanhong Dong. 2018. "Synthesis and Biological Evaluation of Azamacrolide Comprising the Triazole Moiety as Quorum Sensing Inhibitors" Molecules 23, no. 5: 1086. https://doi.org/10.3390/molecules23051086