Theoretical Study of the β-Cyclodextrin Inclusion Complex Formation of Eugenol in Water
Abstract
:1. Introduction
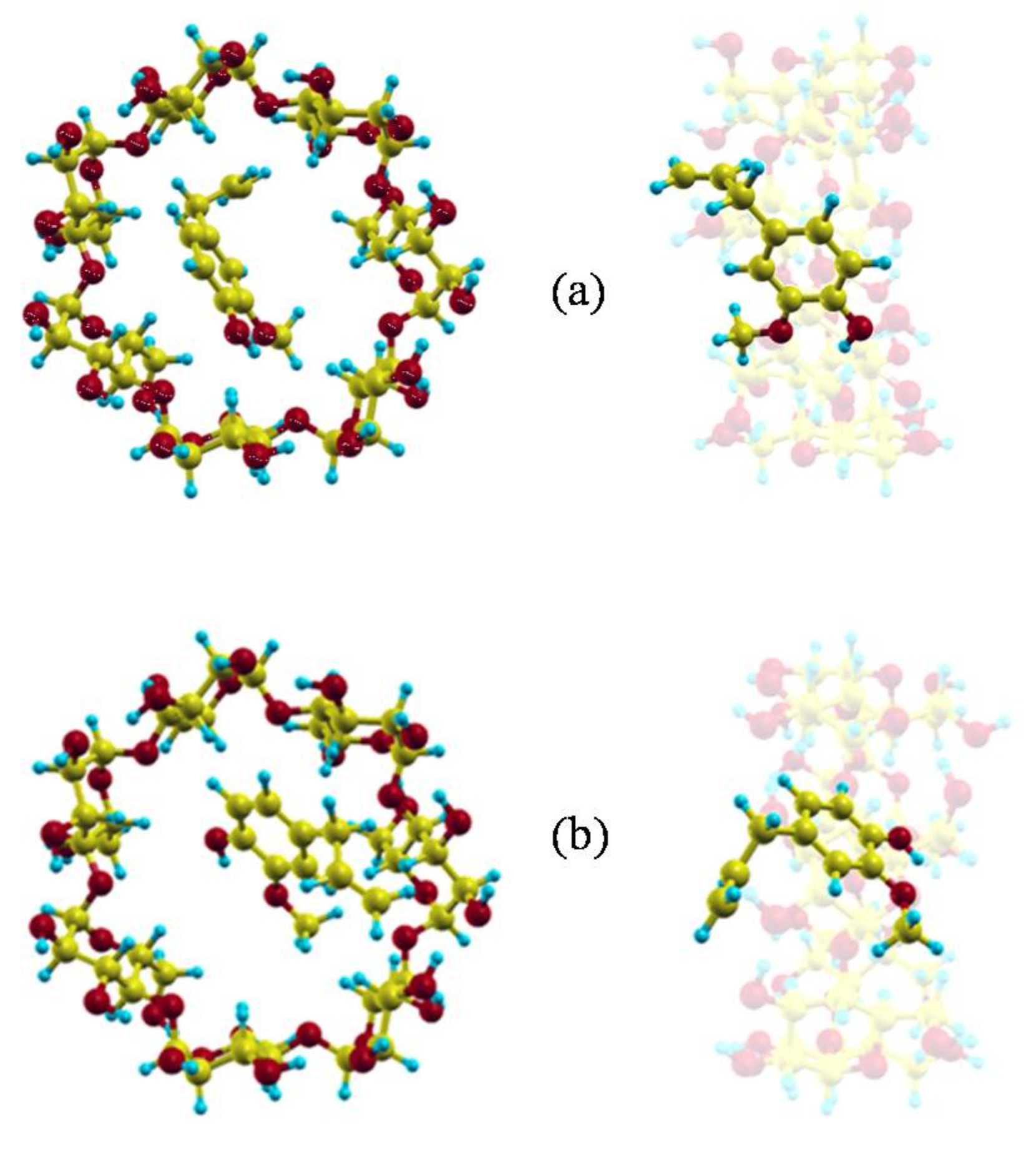
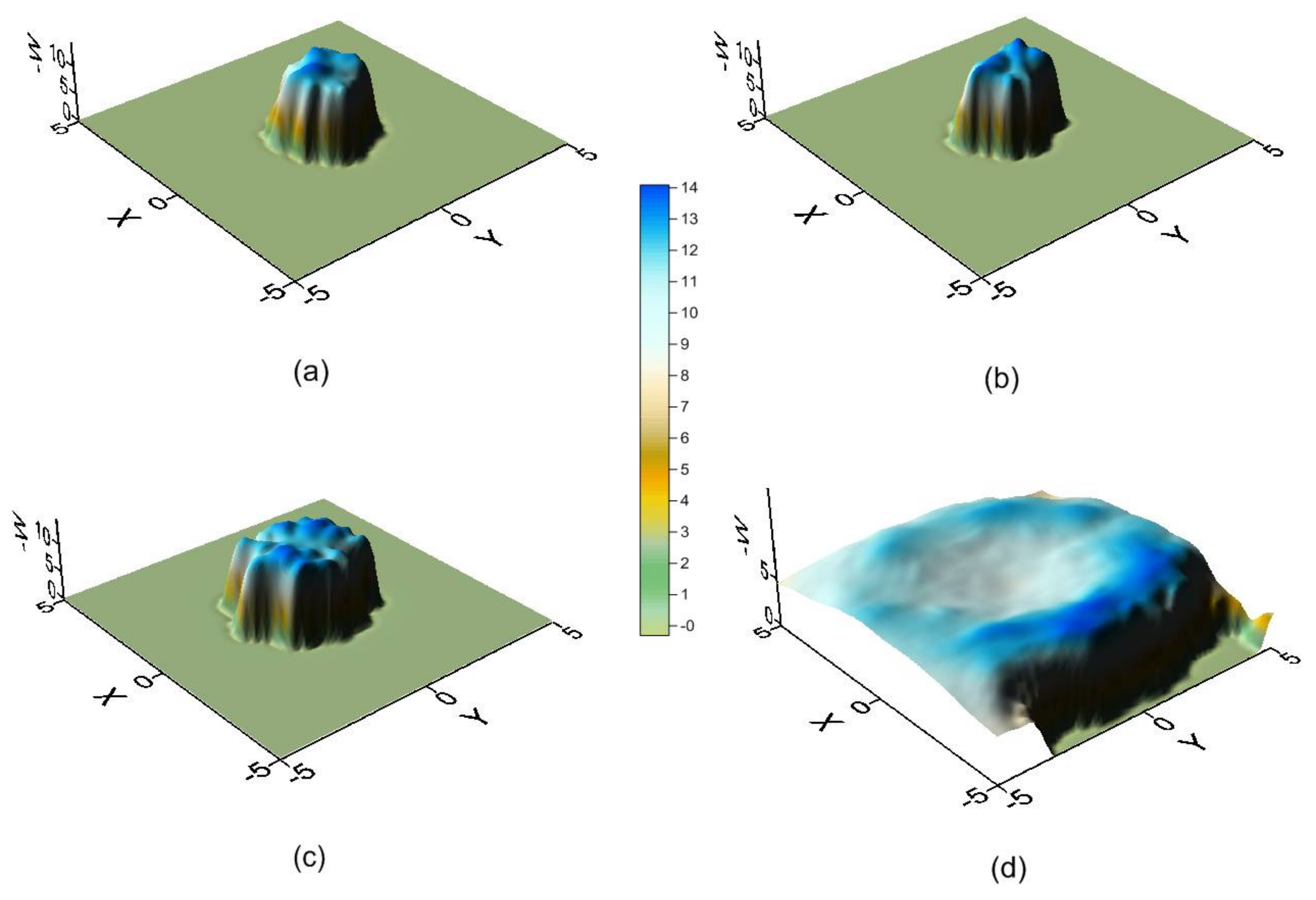
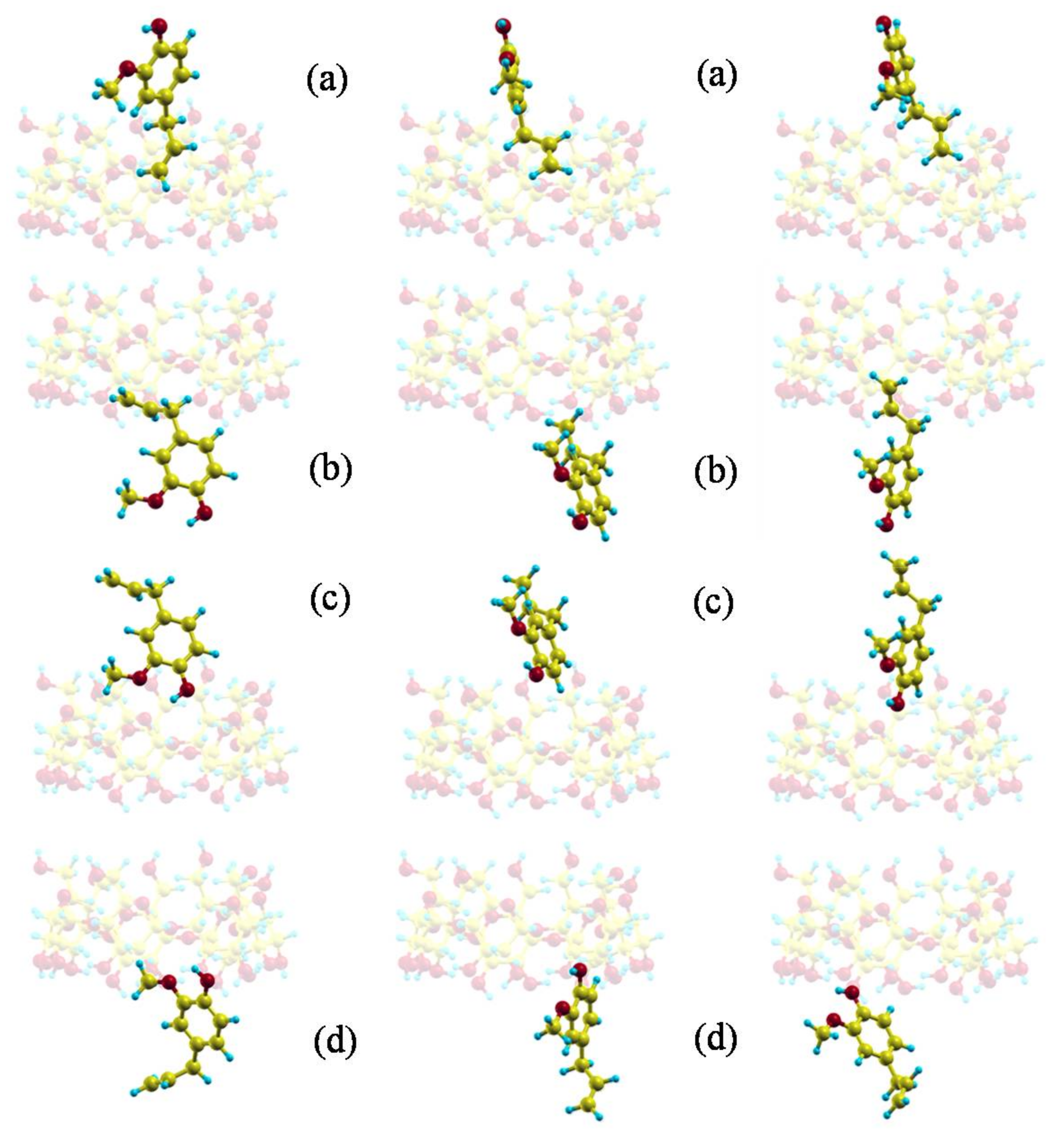
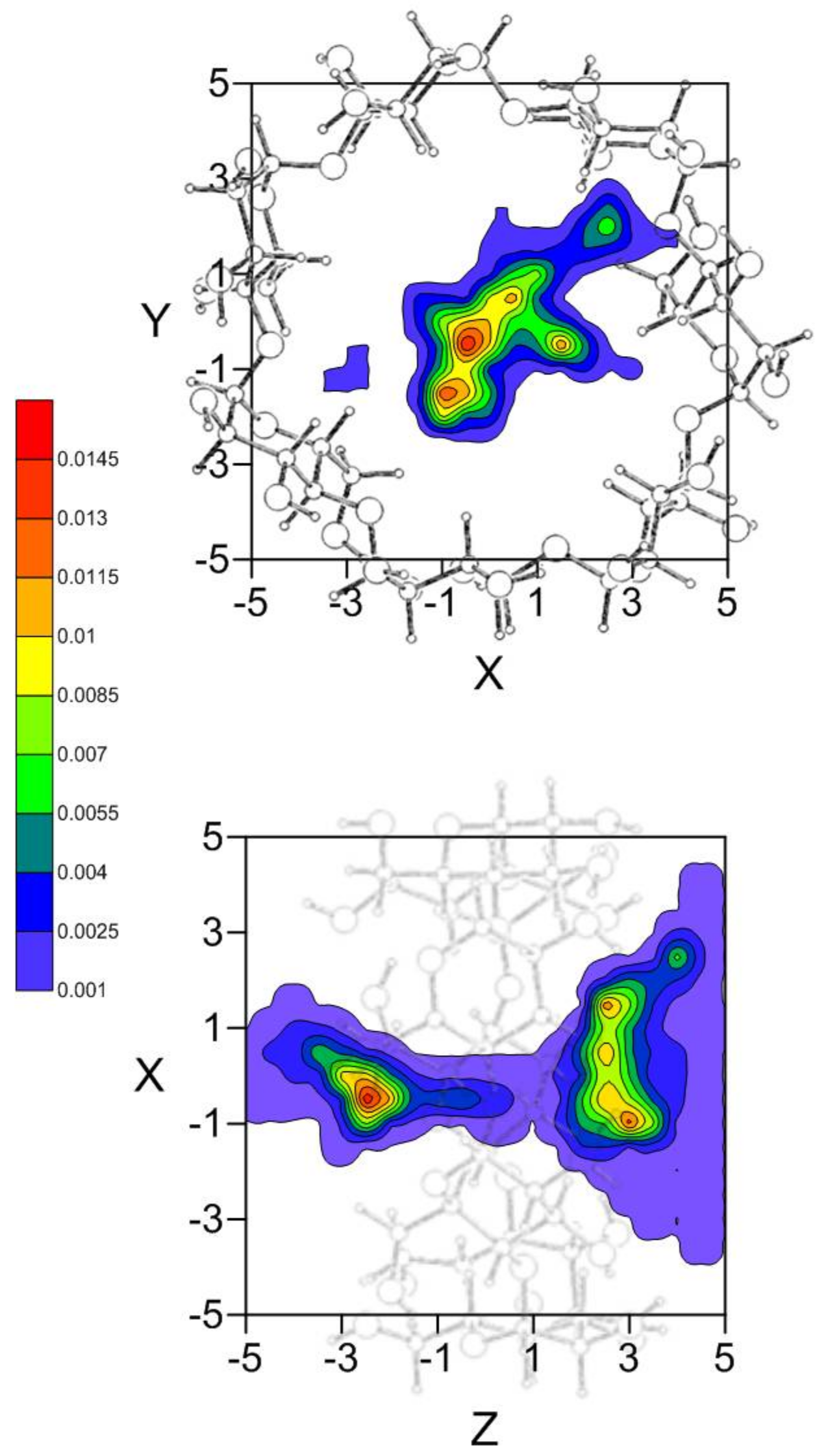
2. Results and Discussion
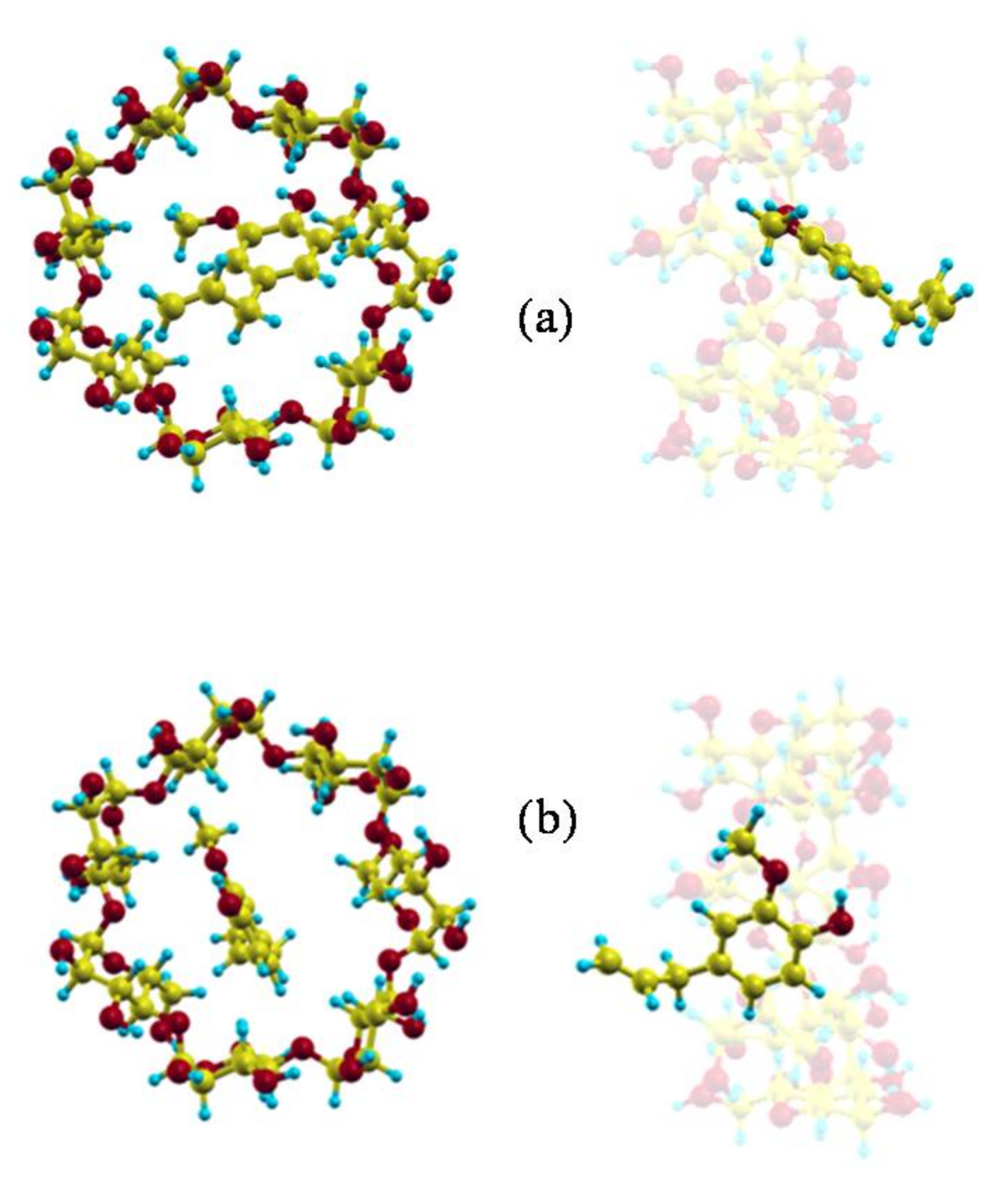
2.1. Molecular Mechanics Simulation
2.2. Molecular Dynamics Simulation
3. Materials and Methods
3.1. Molecular Mechanics Simulation
3.2. Molecular Dynamics Simulation
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Szejtli, J.; Osa, T. Comprehensive Supramolecular Chemistry; Pergamon/Elsevier: Oxford, UK, 1996. [Google Scholar]
- Lipkowitz, K.B. Atomistic modeling of enantioselection in chromatography. J. Chromatogr. A 2001, 906, 417–442. [Google Scholar] [CrossRef]
- Lipkowitz, K.B. Applications of computational chemistry to the study of cyclodextrins. Chem. Rev. 1998, 98, 1829–1873. [Google Scholar] [CrossRef] [PubMed]
- Malta, L.F.B.; Cordeiro, Y.; Tinoco, L.W.; Campos, C.C.; Medeiros, M.E.; Antunes, O.A.C. Recognition mechanism of d- and l-tryptophan enantiomers using 2-hydroxypropyl-α- or β-cyclodextrins as chiral selectors. Tetrahedron Asymmetry 2008, 19, 1182–1188. [Google Scholar] [CrossRef]
- Wang, Y.; Chung, T.S.; Wang, H.; Goh, S.H. Butanol isomer separation using polyamide-imide/CD mixed matrix membranes via pervaporation. Chem. Eng. Sci. 2009, 64, 5198–5209. [Google Scholar] [CrossRef]
- Brocos, P.; Díaz-Vergara, N.; Banquy, X.; Pérez-Casas, S.; Costas, M.; Piñeiro, A. Similarities and differences between cyclodextrin-sodium dodecyl sulfate host-guest complexes of different stoichiometries: Molecular dynamics simulation at several temperatures. J. Phys. Chem. B 2010, 114, 12455–12467. [Google Scholar] [CrossRef] [PubMed]
- González-Gaitano, G.; Sainz-Rojas, P.R.; Isasi, J.R.; Guerrero-Martínez, A.; Tardajos, G. Site-specific interaction between 2-dibenzofuran carboxilate and β- and γ-cyclodextrins determined by intermolecular NOE and molecular modeling. J. Phys. Chem. B 2004, 108, 14154–14162. [Google Scholar] [CrossRef]
- Seridi, L.; Boufelfel, A. Simulations of docking C60 in β-cyclodextrin. J. Mol. Liq. 2011, 162, 69–77. [Google Scholar] [CrossRef]
- Elbashir, A.A.; Suliman, F.E.O. Computational modeling of capillary electrophoretic behavior of primary amines using dual system of 18-crown-6 and β-cyclodextrin. J. Chromatogr. A 2011, 1218, 5344–5351. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Egwolf, B.; Walters, D.E.; Roux, B. Ion selectivity of α-hemolysin with a β-cyclodextrin adapter. I. Single ion potential of mean force and diffusion coefficient. J. Phys. Chem. B 2010, 114, 952–958. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Chipot, C.; Shao, X.; Cai, W. Structural characterization of micelles formed of cholesteryl-functionalized cyclodextrins. Langmuir 2011, 27, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Cairo, P.; Ortuso, F.; Alcaro, S.; Fontananova, E.; Tocci, E.; Drioli, E. β-Cyclodextrin interactions with three drugs used in inflammatory pathologies: An experimental and theoretical study. Chem. Phys. Lett. 2008, 454, 374–381. [Google Scholar] [CrossRef]
- Hernández-Sánchez, P.; López-Miranda, S.; Guardiola, L.; Serrano-Martínez, A.; Gabaldón, J.A.; Núñez-Delicado, E. Optimization of a method for preparing solid complexes of essential clove oil with β-cyclodextrins. J. Sci. Food Agric. 2017, 97, 420–426. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Sánchez, P.; López-Miranda, S.; Lucas-Abellán, C.; Núñez-Delicado, E. Complexation of eugenol (EG), as main component of clove oil and as pure compound, with β- and HP-β-CDs. Food Nutr. Sci. 2012, 3, 716–723. [Google Scholar] [CrossRef]
- Zhan, H.; Jiang, Z.T.; Wang, Y.; Li, R.; Dong, T.S. Molecular microcapsules and inclusion interactions of eugenol with β-cyclodextrin and its derivatives. Eur. Food Res. Technol. 2008, 227, 1507–1513. [Google Scholar] [CrossRef]
- Yang, Y.; Song, L.X. Study on the inclusion compounds of eugenol with α-, β-, γ- and heptakis (2,6-di-O-methyl)-β-cyclodextrins. J. Incl. Phenom. Macrocycl. Chem. 2005, 53, 27–33. [Google Scholar] [CrossRef]
- Divakar, S.; Maheswaran, M.M. Structural studies on inclusion compounds of β-cyclodextrin with some substituted phenols. J. Incl. Phenom. Mol. Recogn. Chem. 1997, 27, 113–126. [Google Scholar] [CrossRef]
- Chowdhry, B.Z.; Ryall, J.P.; Dines, T.J.; Mendham, A.P. Infrared and Raman spectroscopy of eugenol, isoeugenol, and methyl eugenol: Conformational analysis and vibrational assignments from density functional theory calculations of the anharmonic fundamentals. J. Phys. Chem. A 2015, 119, 11280–11292. [Google Scholar] [CrossRef] [PubMed]
- Olbert-Majkut, A.; Wierzejewska, M. Conformational Study of Eugenol by Density Functional Theory Method and Matrix-Isolation Infrared Spectroscopy. J. Phys. Chem. A 2008, 112, 5691–5699. [Google Scholar] [CrossRef] [PubMed]
- Mahboub, R. Structural conformational study of eugenol derivatives using semiempirical methods. Adv. Chem. 2014, 2014, 1–5. [Google Scholar] [CrossRef]
- Sajomsang, W.; Nuchuchua, O.; Gonil, P.; Saesoo, S.; Sramala, I.; Soottitantawat, A.; Puttipipatkhachorn, S.; Ruktanonchai, U.R. Water-soluble β-cyclodextrin grafted with chitosan and its inclusion complex as a mucoadhesive eugenol carrier. Carbohydrate. Polym. 2012, 89, 623–631. [Google Scholar] [CrossRef] [PubMed]
- Alvira, E.; Mayoral, J.A.; Garcia, J.I. Enantiodiscrimination of equol in β-cyclodextrin: An experimental and computational study. J. Incl. Phenom. Macrocycl. Chem. 2008, 60, 103–113. [Google Scholar] [CrossRef]
- Alvira, E. Molecular dynamics study of the influence of solvents on the chiral discrimination of alanine enantiomers by β-cyclodextrin. Tetrahedron Asymmetry 2013, 24, 1198–1206. [Google Scholar] [CrossRef]
- Alvira, E. Theoretical study of the separation of valine enantiomers by β-cyclodextrin with different solvents: A molecular mechanics and dynamics simulation. Tetrahedron Asymmetry 2015, 26, 1198–1206. [Google Scholar] [CrossRef]
- Alvira, E. Influence of solvent polarity on the separation of leucine enantiomers by β-cyclodextrin: A molecular mechanics and dynamics simulation. Tetrahedron Asymmetry 2017, 28, 1414–1422. [Google Scholar] [CrossRef]
- Kokalj, A. Computer graphics and graphical user interfaces as tools in simulations of matter at the atomic scale. Comput. Mater. Sci. 2003, 28, 155–168. [Google Scholar] [CrossRef]
- Liu, L.; Guo, Q.-X. The driving forces in the inclusion complexation of cyclodextrins. J. Incl. Phenom. Macrocycl. Chem. 2002, 42, 1–14. [Google Scholar] [CrossRef]
- Biedermann, F.; Nau, W.M.; Scheneider, H.-J. The hydrophobic effect revisited-studies with supramolecular complexes imply high-energy water as a noncovalent driving force. Angew. Chem. Int. Ed. 2014, 53, 11158–11171. [Google Scholar] [CrossRef] [PubMed]
- Frenkel, D.; Smit, B. Understanding Molecular Simulation; Academic Press: San Diego, CA, USA, 2002. [Google Scholar]
- Rapaport, D.C. The Art of Molecular Dynamics Simulation; Cambridge University Press: Cambridge, UK, 1995. [Google Scholar]
- Kinglert, B.; Rihs, G.J. Molecular encapsulation of transition metal complexes in cyclodextrins. Part 3. Structural consequences of varying the guest geometry in channel-type inclusion compounds. J. Chem. Soc. Dalton Trans. 1991, 2749–2760. [Google Scholar]
- Weiner, S.J.; Kollman, P.A.; Case, D.A.; Singh, U.C.; Ghio, C.; Alagona, G.; Profeta, S., Jr.; Weiner, P.J. A new force field for molecular and mechanical simulation of nucleic acids and proteins. J. Am. Chem. Soc. 1984, 106, 765–784. [Google Scholar] [CrossRef]
- Weiner, S.J.; Kollman, P.A.; Nguyen, D.T.; Case, D.A. An all atom force field for simulations of proteins and nucleic acids. J. Comput. Chem. 1986, 7, 230–252. [Google Scholar] [CrossRef] [PubMed]
- Werner, H.-J.; Knowles, P.J.; Knizia, G.; Manby, F.R.; Schütz, M. Molpro: A general-purpose quantum chemistry program package. WIREs Comput. Mol. Sci. 2012, 2, 242–253. [Google Scholar] [CrossRef]
- Werner, H.-J.; Knowles, P.J.; Knizia, G.; Manby, F.R.; Schütz, M.; Celani, P.; Györffy, W.; Kats, D.; Korona, T.; Lindh, R.; et al. MOLPRO, version 2012.1, a Package of ab initio Programs. Available online: http://www.molpro.net (accessed on 7 November 2017).
- Lipkowitz, K.B.; Pearl, G.; Coner, B.; Peterson, M.A. Explanation of where and how enantioselective binding takes place on permethylated β-cyclodextrin, a chiral stationary phase used in gas chromatography. J. Am. Chem. Soc. 1997, 119, 600–610. [Google Scholar] [CrossRef]
- Lipkowitz, K.B.; Coner, B.; Peterson, M.A. Locating regions of maximum chiral discrimination: A computational study of enantioselection on a popular chiral stationary phase used in chromatography. J Am. Chem. Soc. 1997, 119, 11269–11276. [Google Scholar] [CrossRef]
- Brown, D.; Clarke, J.H.R. A comparison of constant energy, constant temperature and constant pressure ensembles in molecular dynamics simulations of atomic liquids. Mol. Phys. 1984, 51, 1243–1248. [Google Scholar] [CrossRef]
- Fincham, D.; Quirke, N.; Tildesley, D.J. Computer simulation of molecular liquid mixtures. I. A diatomic Lennard-Jones model mixture for CO2/C2H6. J. Chem. Phys. 1986, 84, 4535–4541. [Google Scholar] [CrossRef]
- Izadi, S.; Harris, R.C.; Fenley, M.O.; Onufriev, A.V. Accuracy comparison of generalized Born Models in the calculation of electrostatic binding free energies. J. Chem. Theory Comput. 2018, 14, 1656–1670. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |


© 2018 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alvira, E. Theoretical Study of the β-Cyclodextrin Inclusion Complex Formation of Eugenol in Water. Molecules 2018, 23, 928. https://doi.org/10.3390/molecules23040928
Alvira E. Theoretical Study of the β-Cyclodextrin Inclusion Complex Formation of Eugenol in Water. Molecules. 2018; 23(4):928. https://doi.org/10.3390/molecules23040928
Chicago/Turabian StyleAlvira, Elena. 2018. "Theoretical Study of the β-Cyclodextrin Inclusion Complex Formation of Eugenol in Water" Molecules 23, no. 4: 928. https://doi.org/10.3390/molecules23040928




