Study of Photochemical Cytosine to Uracil Transition via Ultrafast Photo-Cross-Linking Using Vinylcarbazole Derivatives in Duplex DNA
Abstract
:1. Introduction
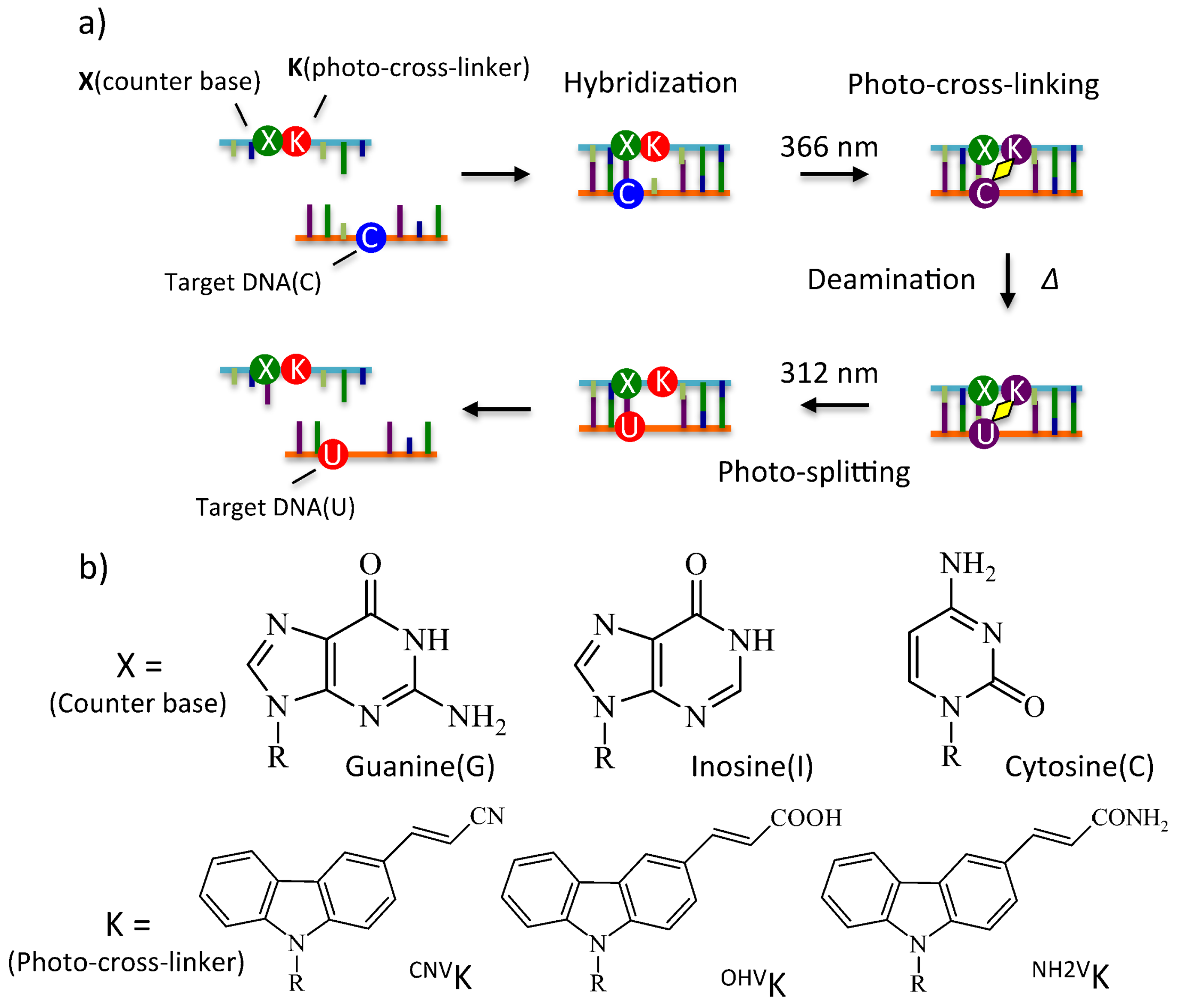
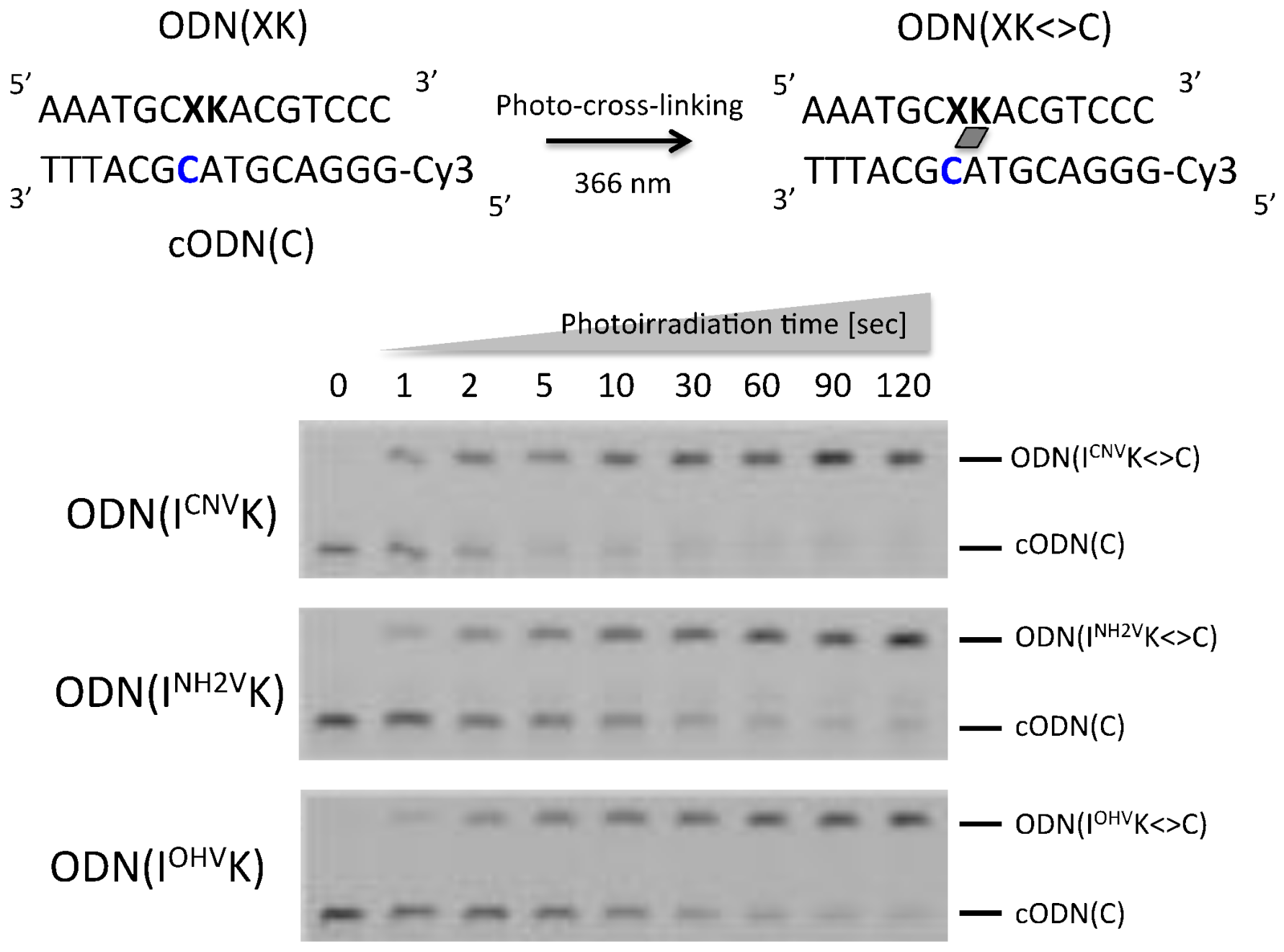
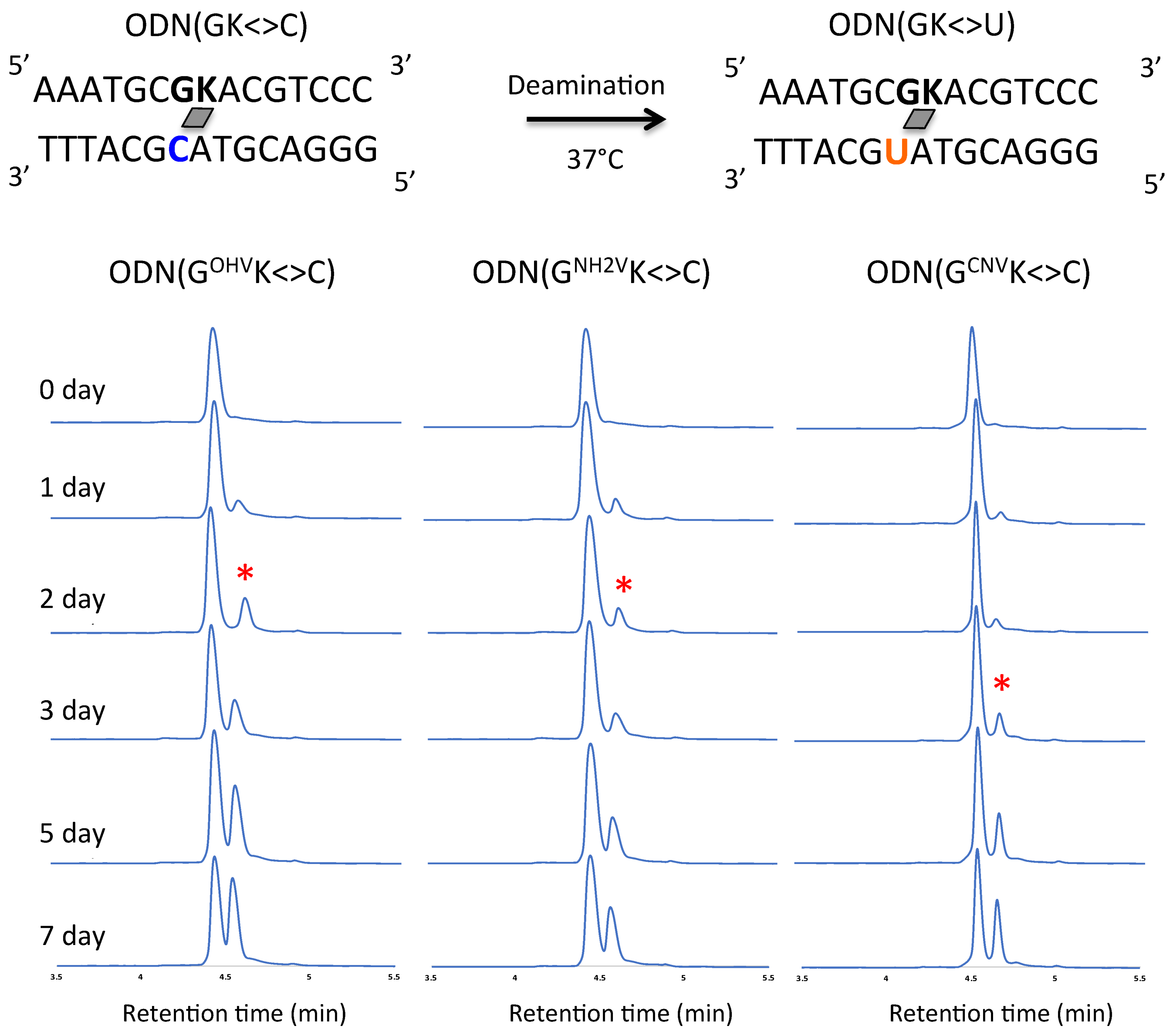
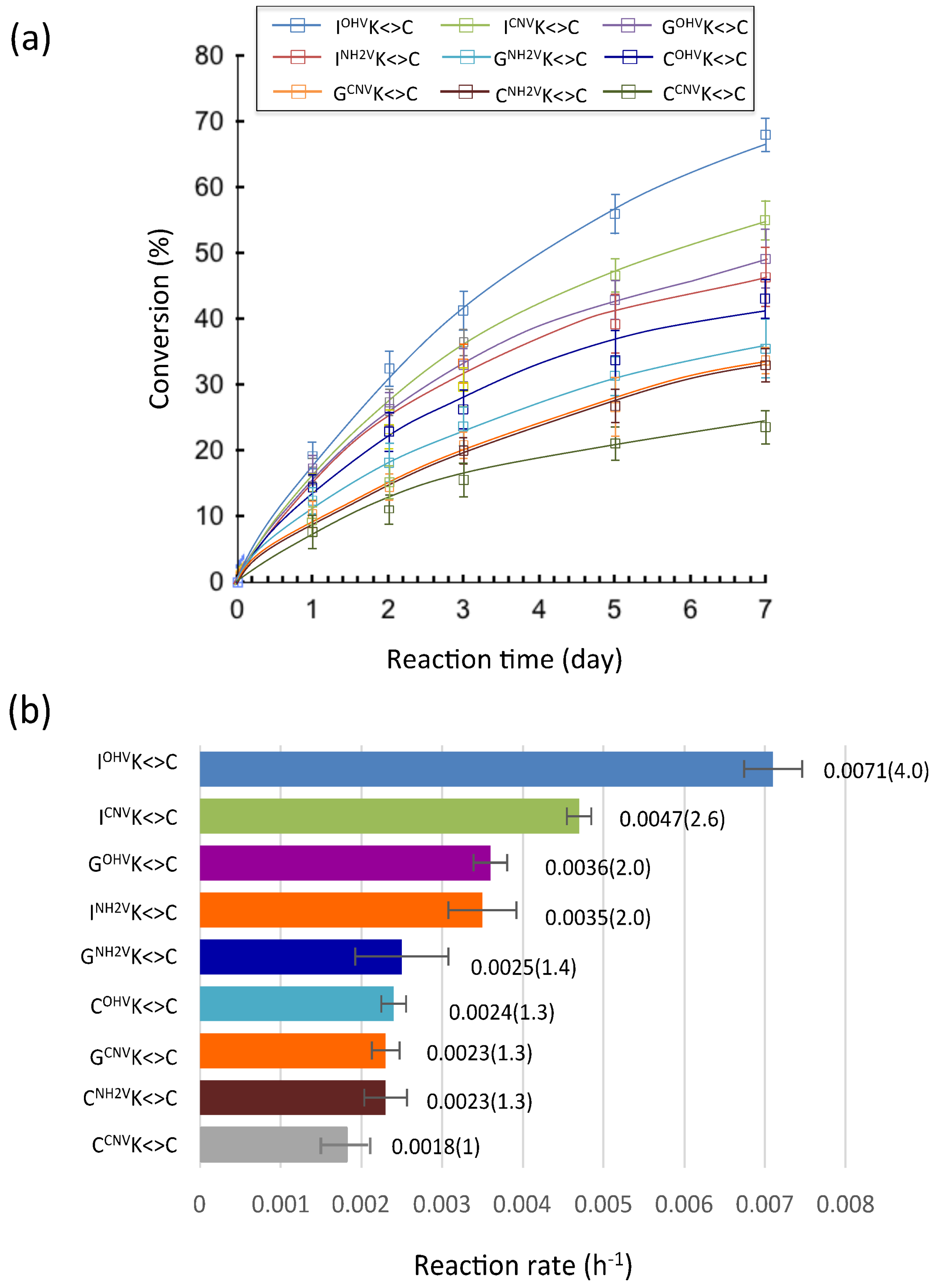
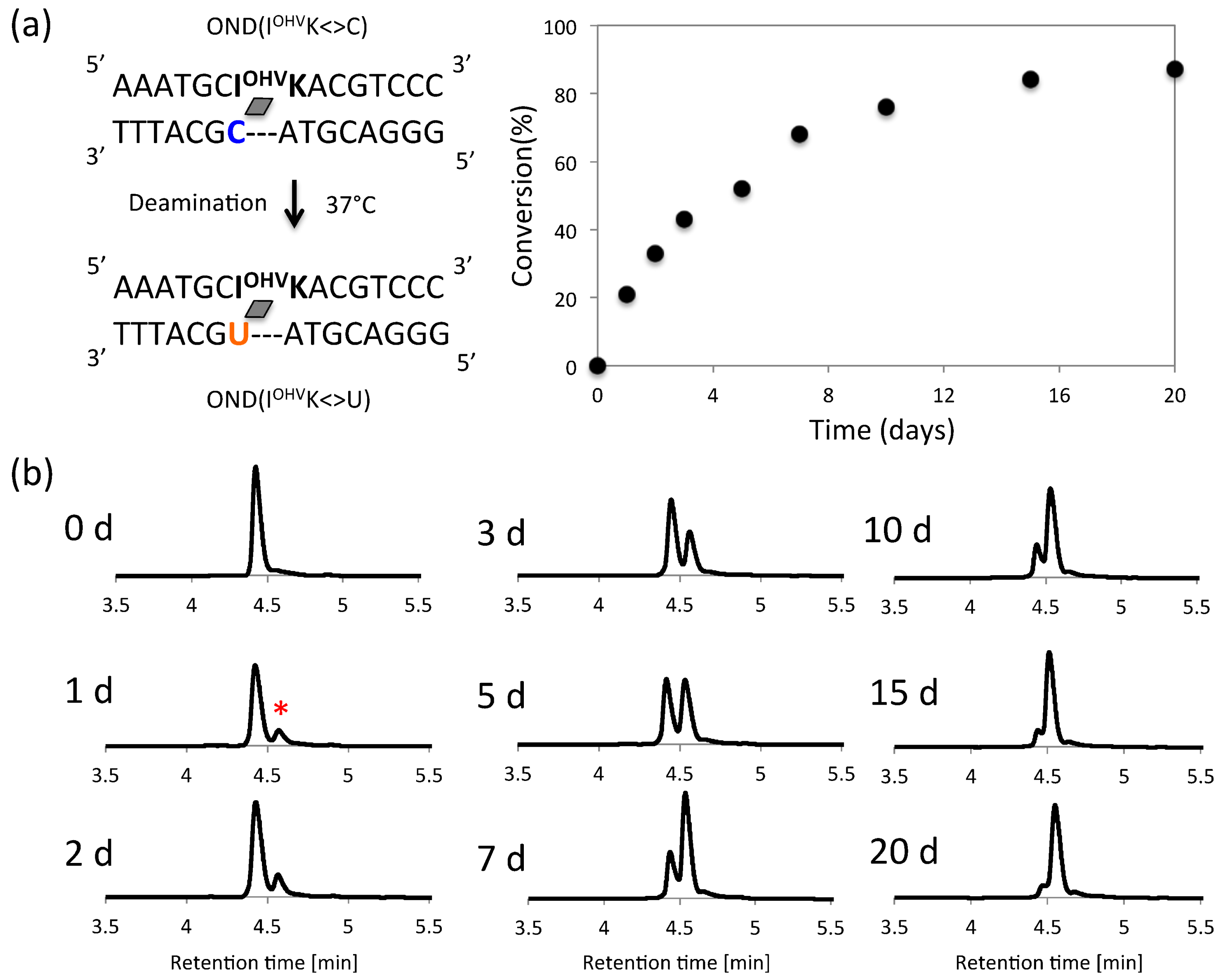
2. Results and Discussion
3. Materials and Methods
3.1. General
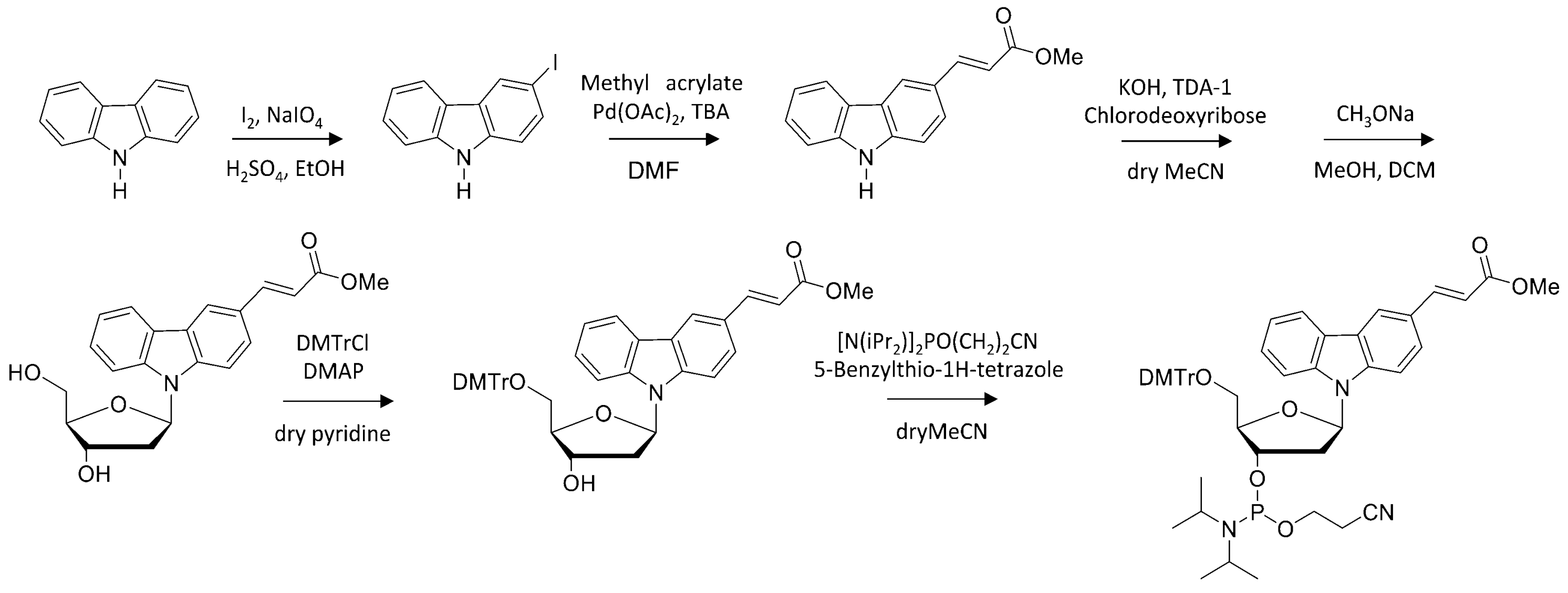
3.2. Synthesis and Preparation of Modified Oligonucleotides
3.3. Denaturing PAGE Analysis
3.4. Isolation of Photo-Cross-Linked dsDNA
3.5. Deamination
3.6. Photo-Splitting and UPLC Analysis
3.7. Partition Coefficient (LogP)
3.8. Enzymatic Digestion
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Cascalho, M. Advantages and disadvantages of cytidine deamination. J. Immunol. 2004, 172, 6513–6518. [Google Scholar] [CrossRef] [PubMed]
- Storici, F.; Lewis, L.K.; Resnick, M.A. In vivo site-directed mutagenesis using oligonucleotides. Nat. Biotechnol. 2001, 19, 773–776. [Google Scholar] [CrossRef] [PubMed]
- Esvelt, K.M.; Wang, H.H. Genome-scale engineering for systems and synthetic biology. Mol. Syst. Biol. 2013, 9, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Tan, W.S.; Carlson, D.F.; Walton, M.W.; Fahrenkrug, S.C.; Hackett, P.B. Precision editing of large animal genomes. Adv. Genet. 2012, 80, 37–97. [Google Scholar] [CrossRef] [PubMed]
- Puchta, H.; Fauser, F. Gene targeting in plants: 25 years later. Int. J. Dev. Biol. 2013, 57, 629–637. [Google Scholar] [CrossRef] [PubMed]
- Sanger, J.D.; Joung, J.K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 2014, 32, 347–355. [Google Scholar] [CrossRef]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-guided human genome engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Bikard, D.; Cox, D.; Zhang, F.; Marraffini, L.A. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 2013, 31, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Javier, N.D.; Michael, N.S. Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature 2002, 419, 43–48. [Google Scholar] [CrossRef]
- White, M.K.; Kaminski, R.; Young, W.B.; Roehm, P.C.; Khalili, K. CRISPR Editing Technology in Biological and Biomedical Investigation. J. Cell. Biochem. 2017, 117, 3586–3594. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.W.; Kim, S.; Kim, J.M.; Kim, J.S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013, 31, 230–232. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.H.; Tee, L.Y.; Wang, X.G.; Huang, Q.S.; Yang, S.H. Off-target Effects in CRISPR/Cas9-mediated Genome Engineering. Mol. Ther.-Nucleic Acids 2015, 4, e264. [Google Scholar] [CrossRef] [PubMed]
- McMahon, M.A.; Rahdar, M.; Porteus, M. Gene editing: Not just for translation anymore. Nat. Methods 2012, 9, 28–31. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.; Grizot, S.; Arnould, S.; Duclert, A.; Epinat, J.C.; Chames, P.; Prieto, J.; Redondo, P.; Blanco, F.J.; Bravo, J.; et al. A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res. 2006, 34, e149. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Kin, J. A guide to genome engineering with programmable nucleases. Nat. Rev. Genet. 2014, 15, 321–334. [Google Scholar] [CrossRef] [PubMed]
- Frommer, M.; McDonald, L.E.; Millar, D.S.; Collis, C.M.; Watt, F.; Grigg, G.W.; Molloy, P.L.; Paul, C.L. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. USA 1992, 89, 1827–1831. [Google Scholar] [CrossRef] [PubMed]
- Yamana, K.; Yoshikawa, A.; Nakano, H. Synthesis of a new photoisomerizable linker for connecting two oligonucleotide segments. Tetrahedron Lett. 1996, 37, 637–640. [Google Scholar] [CrossRef]
- Lee, B.L.; Blake, K.R.; Miller, P.S. Interaction of psoralen-derivatized oligodeoxyribonucleoside methylphosphonates with synthetic DNA containing a promoter for T7 RNA polymerase. Nucleic Acids Res. 1988, 16, 10681–10697. [Google Scholar] [CrossRef] [PubMed]
- Montes, C.V.; Memczak, H.; Gyssels, E.; Torres, T.; Madder, A.; Schneider, R.J. Photoinduced Cross-Linking of Short Furan-Modified DNA on Surfaces. Langmuir 2017, 33, 1197–1201. [Google Scholar] [CrossRef] [PubMed]
- Kurz, M.; Gu, K.; Lohse, P.A. Psoralen photo-crosslinked mRNA-puromycin conjugates: A novel template for the rapid and facile preparation of mRNA-protein fusions. Nucleic Acids Res. 2000, 28, 83. [Google Scholar] [CrossRef]
- Liang, X.; Wakuda, R.; Fujioka, K.; Asanuma, H. Photoregulation of DNA transcription by using photoresponsive T7 promoters and clarification of its mechanism. FEBS J. 2010, 277, 1551–1561. [Google Scholar] [CrossRef]
- Liu, J.; Geng, Y.; Pound, E.; Gyawall, S.; Ashton, J.R.; Hickey, J.; Woolley, A.T.; Harb, J.N. Metallization of branched DNA origami for nanoelectronic circuit fabrication. ACS Nano 2011, 5, 2240–2247. [Google Scholar] [CrossRef] [PubMed]
- Iwase, R.; Namba, M.; Yamaoka, T.; Murakami, A. Gene regulation by decoy approach (I): Synthesis and properties of photo-crosslinked oligonucleotides. Nucleic Acids Symp. Ser. 1997, 37, 203–204. [Google Scholar]
- Fujimoto, K.; Konishi-Hiratsuka, K.; Sakamoto, T.; Yoshimura, Y. Site-specific photochemical RNA editing. Chem. Commun. 2010, 46, 7545–7547. [Google Scholar] [CrossRef]
- Fujimoto, K.; Konishi-Hiratsuka, K.; Sakamoto, T.; Yoshimura, Y. Site-Specific Cytosine to Uracil Transition by Using Reversible DNA Photo-crosslinking. ChemBioChem 2010, 11, 1661–1664. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, T.; Ooe, M.; Fujimoto, K. Critical Effect of Base Pairing of Target Pyrimidine on the Interstrand Photo-Cross-Linking of DNA via 3-Cyanovinylcarbazole Nucleoside. Bioconj. Chem. 2015, 26, 1475–1478. [Google Scholar] [CrossRef] [PubMed]
- Sethi, S.; Ooe, M.; Sakamoto, T.; Fujimoto, K. Effect of nucleobase change on cytosine deamination through DNA photo-cross-linking reaction via 3-cyanovinylcarbazole nucleoside. Mol. BioSyst. 2017, 13, 1152–1156. [Google Scholar] [CrossRef] [PubMed]
- Sethi, S.; Yasuharu, T.; Nakamura, S.; Fujimoto, K. Effect of substitution of photo-cross-linker in photochemical cytosine to uracil transition in DNA. Bioorg. Med. Chem. Lett. 2017, 27, 3905–3908. [Google Scholar] [CrossRef] [PubMed]
- OECD Guidline for Testing of Chemicals. Available online: http://www.oecd.org/chemicalsafety/risk-assessment/1948177.pdf (accessed on 15 December 2017).
- Borges, N.M.; Kenny, P.W.; Montanari, C.A.; Prokopozyk, I.M.; Ribeiro, J.F.; Rocha, J.R.; Sartori, G.R. The influence of hydrogen bonding on partition coefficients. J. Comput.-Aided Mol. Des. 2017, 31, 163–181. [Google Scholar] [CrossRef] [PubMed]
- Sklenak, S.; Yao, L.; Cukier, R.I.; Yan, H. Catalytic mechanism of yeast cytosine deaminase: An ONIOM computational study. J. Am. Chem. Soc. 2004, 126, 14879–14889. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhao, Y.; Yan, H.; Cao, Z. Combined QM(DFT)/MM molecular dynamics simulations of the deamination of cytosine by yeast cytosine deaminase (yCD). J. Comput. Chem. 2016, 37, 1163–1174. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, Y.; Fujimoto, K. Ultrafast reversible photo-cross-linking reaction: Toward in situ DNA manipulation. Org. Lett. 2008, 10, 3227–3230. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are available from the authors. |



| Entry | Sequence of ODNs (5′ to 3′) |
|---|---|
| ODN(GCNVK) | AAATGCGCNVKACGTCCC |
| ODN(GNH2VK) | AAATGCGNH2VKACGTCCC |
| ODN(GOHVK) | AAATGCGOHVKACGTCCC |
| ODN(CCNVK) | AAATGCCCNVKACGTCCC |
| ODN(CNH2VK) | AAATGCCNH2VKACGTCCC |
| ODN(COHVK) | AAATGCCOHVKACGTCCC |
| ODN(ICNVK) | AAATGCICNVKACGTCCC |
| ODN(INH2VK) | AAATGCINH2VKACGTCCC |
| ODN(IOHVK) | AAATGCIOHVKACGTCCC |
| Entry | Retention Time (min) | LogP | |
|---|---|---|---|
| ODN(IOHVK<>C) | 13.8 | 0.52 | |
| ODN(GOHVK<>C) | 14.2 | 0.54 | |
| ODN(COHVK<>C) | 14.7 | 0.57 | |
| ODN(INH2VK<>C) | 15.4 | 0.61 | |
| ODN(GNH2VK<>C) | 16.0 | 0.64 | |
| ODN(ICNVK<>C) | 16.3 | 0.66 | |
| ODN(GCNVK<>C) | 16.6 | 0.68 | |
| ODN(CCNVK<>C) | 17.3 | 0.72 | |
| ODN(CNH2VK<>C) | 17.8 | 0.75 | |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sethi, S.; Nakamura, S.; Fujimoto, K. Study of Photochemical Cytosine to Uracil Transition via Ultrafast Photo-Cross-Linking Using Vinylcarbazole Derivatives in Duplex DNA. Molecules 2018, 23, 828. https://doi.org/10.3390/molecules23040828
Sethi S, Nakamura S, Fujimoto K. Study of Photochemical Cytosine to Uracil Transition via Ultrafast Photo-Cross-Linking Using Vinylcarbazole Derivatives in Duplex DNA. Molecules. 2018; 23(4):828. https://doi.org/10.3390/molecules23040828
Chicago/Turabian StyleSethi, Siddhant, Shigetaka Nakamura, and Kenzo Fujimoto. 2018. "Study of Photochemical Cytosine to Uracil Transition via Ultrafast Photo-Cross-Linking Using Vinylcarbazole Derivatives in Duplex DNA" Molecules 23, no. 4: 828. https://doi.org/10.3390/molecules23040828






