MiR200 and miR302: Two Big Families Influencing Stem Cell Behavior †
Abstract
:1. Introduction
2. Stem Cells
- Asymmetric divisions with the formation of stem cells (self-regeneration or self-renewal) and daughter cells with reduced differentiation potential, which transiently replicate and thus differentiate after a specific number of divisions;
- The persistence of the replicative capacity for the entire life of the individual;
- The maintenance a source of stem cells due to a specific microenvironment (stem cell niche) formed by other cells.
3. Embryonic and Adult Stem Cells. Cellular Reprogramming and Generation of Pluripotent Stem Cells
4. Induced Pluripotent Stem Cells (iPSCs)
5. miRNAs
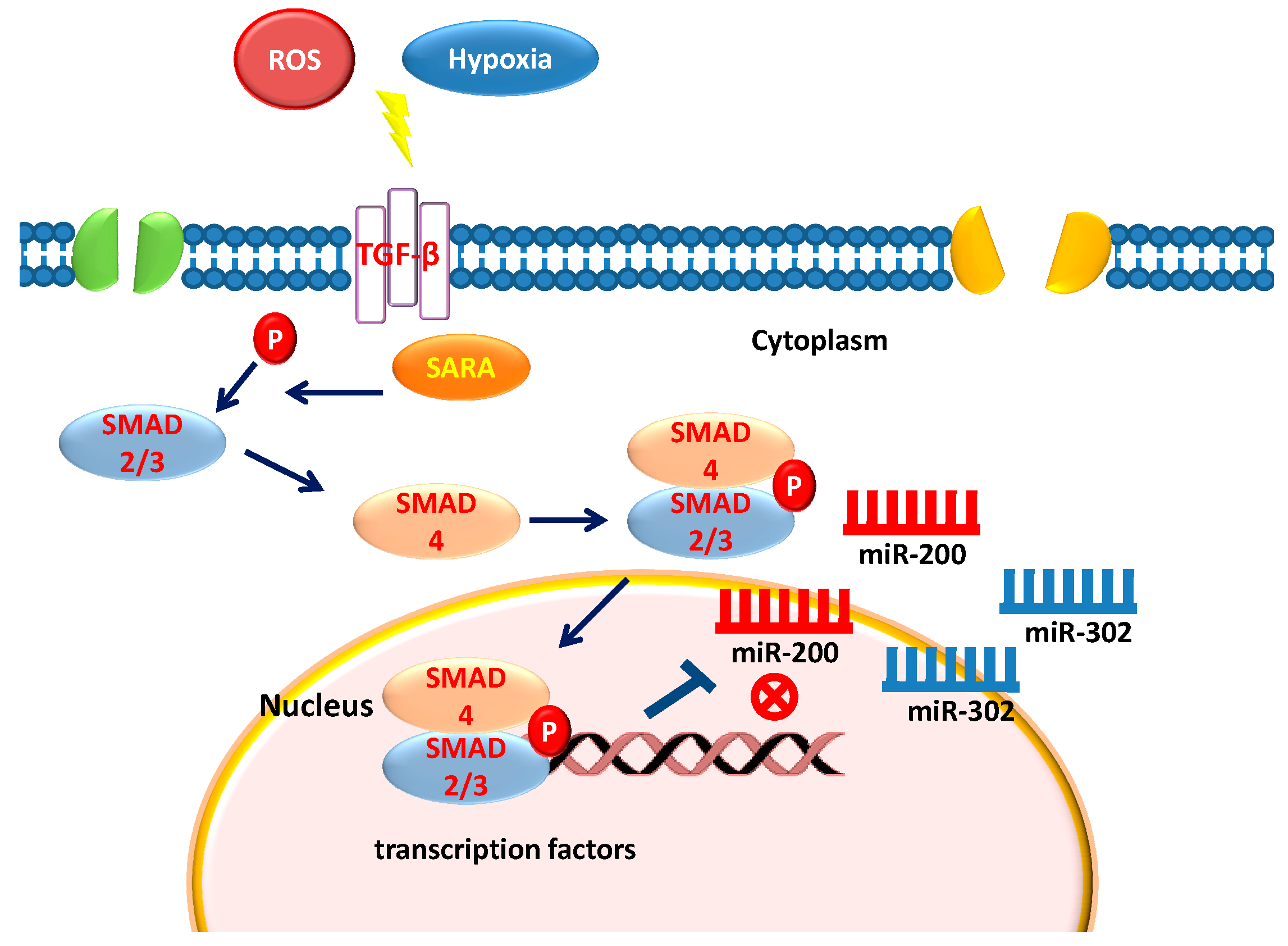
6. Stem Cells, TGF-β, and Epigenetics
7. MicroRNAs and Cell Reprogramming
8. Stem Cell Function and miRNA Transcription
9. MiRNAs Orchestrating Stem Cell Pluripotency and Differentiation
10. The Two Big miRNA Families of Pluripotency: miR-200 and miR-302/367
10.1. MiRNA-200 Family
10.2. The miR-302/367 Family
11. Epithelial–Mesenchymal Transition (EMT) in Stem Cells
12. Hypoxia Exerts Opposite Effect on miR-200 and miR-302 Family
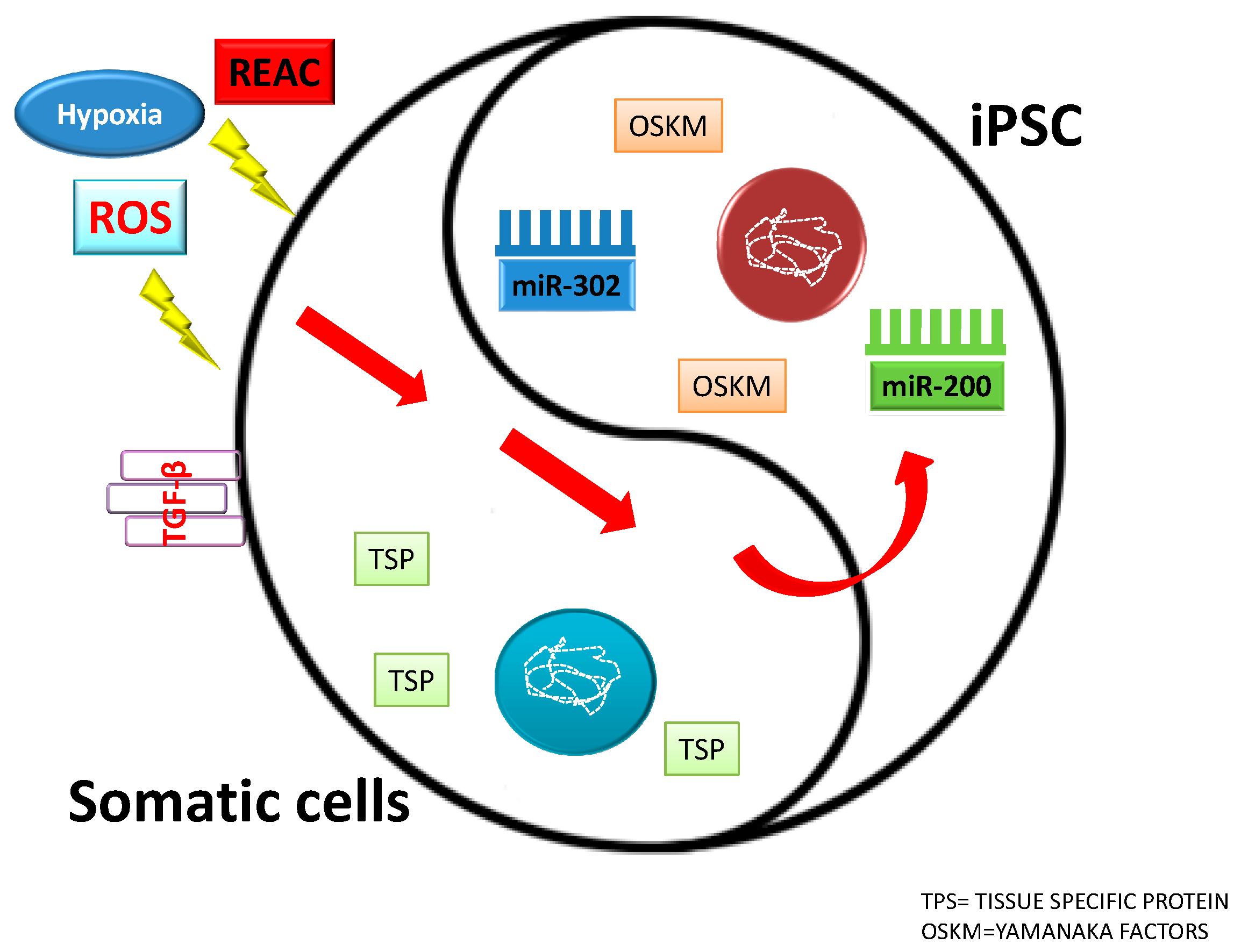
13. Physical Stimuli: Ad Hoc Conveyance of Electromagnetic Fields to Drive Stem Cell Fate
14. Electromagnetic Fields
15. ROS and miRNA
16. Conclusions
Acknowledgments
Conflicts of Interest
References
- Dupont, C.; Armant, D.R.; Brenner, C.A. Epigenetics: Definition, mechanisms and clinical perspective. Semin. Reprod. Med. 2009, 27, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Marson, A.; Levine, S.S.; Cole, M.F.; Frampton, G.M.; Brambrink, T.; Johnstone, S.; Guenther, M.G.; Johnston, W.K.; Wernig, M.; Newman, J.; et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 2008, 134, 521–533. [Google Scholar] [CrossRef] [PubMed]
- Houbaviy, H.B.; Murray, M.F.; Sharp, P.A. Embryonic stem cell-specific MicroRNAs. Dev. Cell 2003, 5, 351–358. [Google Scholar] [CrossRef]
- Bar, M.; Wyman, S.K.; Fritz, B.R.; Qi, J.; Garg, K.S.; Parkin, R.K.; Kroh, E.M.; Bendoraite, A.; Mitchell, P.S.; Nelson, A.M.; et al. MicroRNA discovery and profiling in human embryonic stem cells by deep sequencing of small RNA libraries. Stem Cells 2008, 26, 2496–2505. [Google Scholar] [CrossRef] [PubMed]
- Morin, R.D.; O’Connor, M.D.; Griffith, M.; Kuchenbauer, F.; Delaney, A.; Prabhu, A.L.; Zhao, Y.; McDonald, H.; Zeng, T.; Hirst, M.; et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008, 18, 610–621. [Google Scholar] [CrossRef] [PubMed]
- Selbach, M.; Schwanhausser, B.; Thierfelder, N.; Fang, Z.; Khanin, R.; Rajewsky, N. Widespread changes in protein synthesis induced by microRNAs. Nature 2008, 455, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Berezikov, E.; Guryev, V.; van de Belt, J.; Wienholds, E.; Plasterk, R.H.; Cuppen, E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell 2005, 120, 21–24. [Google Scholar] [CrossRef] [PubMed]
- Blahna, M.T.; Hata, A. Smad-mediated regulation of microRNA biosynthesis. FEBS Lett. 2012, 586, 1906–1912. [Google Scholar] [CrossRef] [PubMed]
- Elston, R.; Inman, G.J. Crosstalk between p53 and tgf-beta signaling. J. Signal Transduct. 2012, 2012, 294097. [Google Scholar] [PubMed]
- Chang, K.C.; Wang, C.; Wang, H. Balancing self-renewal and differentiation by asymmetric division: Insights from brain tumor suppressors in Drosophila neural stem cells. Bioessays 2012, 34, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Shen, D.; Shen, J.; Gao, S.M.; Li, B.; Wong, C.; Feng, W.; Song, Y. The Super Elongation Complex Drives Neural Stem Cell Fate Commitment. Dev. Cell 2017, 40, 537–551. [Google Scholar] [CrossRef] [PubMed]
- Thomson, J.A.; Itskovitz-Eldor, J.; Shapiro, S.S.; Waknitz, M.A.; Swiergiel, J.J.; Marshall, V.S.; Jones, J.M. Embryonic stem cell lines derived from human blastocysts. Science 1998, 282, 1145–1147. [Google Scholar] [CrossRef] [PubMed]
- Hirai, H.; Karian, P.; Kikyo, N. Regulation of embryonic stem cell self-renewal and pluripotency by leukaemia inhibitory factor. Biochem. J. 2011, 438, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Bi, Y.; Gao, S. Epigenetic regulation of somatic cell reprogramming. Curr. Opin. Genet. Dev. 2017, 46, 156–163. [Google Scholar] [CrossRef] [PubMed]
- Butz, H.; Rácz, K.; Hunyady, L.; Patócs, A. Crosstalk between TGF-β signaling and the microRNA machinery. Trends Pharmacol. Sci. 2012, 33, 382–393. [Google Scholar] [CrossRef] [PubMed]
- Tay, Y.; Zhang, J.; Thomson, A.M.; Lim, B.; Rigoutsos, I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature 2008, 455, 1124–1128. [Google Scholar] [CrossRef] [PubMed]
- Creemers, E.E.; Tijsen, A.J.; Pinto, Y.M. Circulating microRNAs: Novel biomarkers and extracellular communicators in cardiovascular disease? Circ. Res. 2012, 110, 483–495. [Google Scholar] [CrossRef] [PubMed]
- Bar-Nur, O.; Russ, H.A.; Efrat, S.; Benvenisty, N. Epigenetic memory and preferential lineage-specific differentiation in induced pluripotent stemcells derived from human pancreatic islet beta cells. Cell Stem Cell 2011, 9, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, L.; Tilgner, K.; Saretzki, G.; Atkinson, S.P.; Stojkovic, M.; Moreno, R.; Przyborski, S.; Lako, M. Human induced pluripotent stem cell lines show stress defense mechanisms and mitochondrial regulation similar to those of human embryonic stem cells. Stem Cells 2010, 28, 661–673. [Google Scholar] [CrossRef] [PubMed]
- Prigione, A.; Fauler, B.; Lurz, R.; Lehrach, H.; Adjaye, J. The senescence-related mitochondrial/oxidative stress pathway is repressed in human induced pluripotent stem cells. Stem Cells 2010, 28, 721–733. [Google Scholar] [CrossRef] [PubMed]
- Folmes, C.D.; Nelson, T.J.; Martinez-Fernandez, A.; Arrell, D.K.; Lindor, J.Z.; Dzeja, P.P.; Ikeda, Y.; Perez-Terzic, C.; Terzic, A. Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metab. 2011, 14, 264–271. [Google Scholar] [CrossRef] [PubMed]
- Folmes, C.D.; Dzeja, P.P.; Nelson, T.J.; Terzic, A. Metabolic plasticity in stem cell homeostasis and differentiation. Cell Stem Cell 2012, 11, 596–606. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Nuebel, E.; Daley, G.Q.; Koehler, C.M.; Teitell, M.A. Metabolic regulation in pluripotent stem cells during reprogramming and self-renewal. Cell Stem Cell 2012, 11, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Berardi, E.; Pues, M.; Thorrez, L.; Sampaolesi, M. miRNAs in ESC differentiation. Am. J. Physiol. Heart Circ. Physiol. 2012, 15, 931–939. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, A.; Griffiths-Jones, S.; Ashurst, J.L.; Bradley, A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004, 14, 1902–1910. [Google Scholar] [CrossRef] [PubMed]
- Davis-Dusenbery, B.N.; Hata, A. Mechanisms of control of microRNA biogenesis. J. Biochem. 2010, 148, 381–392. [Google Scholar] [PubMed]
- Baek, D.; Villén, J.; Shin, C.; Camargo, F.D.; Gygi, S.P.; Bartel, D.P. The impact of microRNAs on protein output. Nature 2008, 455, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Lüningschrö, P.; Hauser, S.; Kaltschmidtab, B.; Kaltschmidta, C. MicroRNAs in pluripotency, reprogramming and cell fate induction. BBA-Mol. Cell Res. 2013, 8, 1894–1903. [Google Scholar]
- Min, P.K.; Chan, S.Y. The biology of circulating microRNAs in cardiovascular disease. Eur. J. Clin. Investig. 2015, 45, 860–874. [Google Scholar] [CrossRef] [PubMed]
- Maioli, M.; Santaniello, S.; Montella, A.; Bandiera, P.; Cantoni, S.; Bianchi, C.C.F.; Lionetti, V.; Rizzolio, F.; Marchesi, I.; Bagella, L.; et al. Hyaluronan esters drive Smad gene expression and signaling enhancing cardiogenesis in mouse embryonic and human mesenchymal stem cells. PLoS ONE 2010, 5, e15151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Massagué, J. TGFβ signaling in context. Nat. Rev. Mol. Cell Biol. 2012, 13, 616–630. [Google Scholar] [CrossRef] [PubMed]
- Nakao, A.; Afrakhte, M.; Morén, A.; Nakayama, T.; Christian, J.L.; Heuchel, R.; Itoh, S.; Kawabata, M.; Heldin, N.E.; Heldin, C.H.; et al. Identification of Smad7, a TGFbeta-inducible antagonist of TGF-beta signaling. Nature 1997, 389, 631–635. [Google Scholar] [PubMed]
- Massagué, J.; Blain, S.W.; Lo, R.S. TGFbeta signaling in growth control, cancer, and heritable disorders. Cell 2000, 103, 295–309. [Google Scholar] [CrossRef]
- Sakaki-Yumoto, M.; Katsuno, Y.; Derynck, R. TGF-β family signaling in stem cells. Biochim. Biophys. Acta 2013, 1830, 2280–2296. [Google Scholar] [CrossRef] [PubMed]
- Pera, M.F.; Tam, P.P.L. Extrinsic regulation of pluripotent stem cells. Nature 2010, 465, 713–720. [Google Scholar] [CrossRef] [PubMed]
- Warmflash, A.; Arduini, B.L.; Brivanlou, A.H. The molecular circuitry underlying pluripotency in embryonic stem cells. Wiley Interdiscip. Rev. Syst. Biol. Med. 2012, 4, 443–456. [Google Scholar] [CrossRef] [PubMed]
- Boyer, L.A.; Lee, T.I.; Cole, M.F.; Johnstone, S.E.; Levine, S.S.; Zucker, J.P.; Guenther, M.G.; Kumar, R.M.; Murray, H.L.; Jenner, R.G.; et al. Core Transcriptional Regulatory Circuitry in Human Embryonic Stem Cells. Cell 2005, 122, 947–956. [Google Scholar] [CrossRef] [PubMed]
- Vallier, L.; Mendjan, S.; Brown, S.; Chng, Z.; Teo, A.; Smithers, L.E.; Trotter, M.W.B.; Cho, C.H.; Martinez, A.; Rugg-Gunn, P.; et al. Activin/Nodal signaling maintains pluripotency by controlling Nanog expression. Development 2009, 136, 1339–1349. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.M.; Reynolds, D.; Cliff, T.; Ohtsuka, S.; Mattheyses, A.L.; Sun, Y.; Menendez, L.; Kulik, M.; Dalton, S. Signaling Network Crosstalk in Human Pluripotent Cells: A Smad2/3-Regulated Switch that Controls the Balance between Self-Renewal and Differentiation. Cell Stem Cell 2012, 10, 312–326. [Google Scholar] [CrossRef] [PubMed]
- Fedele, M.; Cerchia, L.; Chiappetta, G. The Epithelial-to-Mesenchymal Transition in Breast Cancer: Focus on Basal-Like Carcinomas. Cancers 2017, 9, 134. [Google Scholar] [CrossRef] [PubMed]
- Lima, J.F.; Nofech-Mozes, S.; Bayani, J.; Bartlett, J.M. EMT in breast carcinoma—A Review. J. Clin. Med. 2016, 5, 65. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 12, 663–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sridharan, N.M.R.; Xie, W.; Utikal, J.; Eminli, S.; Arnold, K.; Stadtfeld, M.; Yachechko, R.; Tchieu, J.; Jaenisch, R.; Plath, K.; et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell 2007, 1, 55–70. [Google Scholar]
- Guo, L.; Zhang, Y.; Zhang, L.; Huang, F.; Li, J.; Wang, S. MicroRNAs, TGF-β signaling, and the inflammatory microenvironment in cancer. Tumor Biol. 2016, 37, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Xu, N.; Papagiannakopoulos, T.; Pan, G.; Thomson, J.A.; Kosik, K.S. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell 2009, 137, 647–658. [Google Scholar] [CrossRef] [PubMed]
- Wellner, U.; Schubert, J.; Burk, U.C.; Schmalhofer, O.; Zhu, F.; Sonntag, A.; Waldvogel, B.; Vannier, C.; Darling, D.; zur Hausen, A.; et al. The EMT-activator ZEB1 promotes tum-origenicity by repressing stemness-inhibiting microRNAs. Nat. Cell Biol. 2009, 11, 1487–1495. [Google Scholar] [CrossRef] [PubMed]
- Triboulet, R.; Chang, H.; LaPierre, R.J.; Gregory, R.I. Post-transcriptional control of DGCR8 expression by the Microprocessor. RNA 2009, 15, 1005–1011. [Google Scholar] [CrossRef] [PubMed]
- Annes, J.P.; Munger, J.S.; Rifkin, D.B. Making sense of latent TGFβ activation. J. Cell Sci. 2003, 116, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Dijke, P.T.; Hill, C.S. New insights into TGF-β-Smadsignalling. Trends Biochem. Sci. 2004, 29, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Richter, K.; Kietzmann, T. Reactive oxygen species and fibrosis: Further evidence of a significant liaison. Cell Tissue Res. 2016, 365, 591–605. [Google Scholar] [CrossRef] [PubMed]
- Massagué, J.; Gomis, R.R. The logic of TGFβ signaling. FEBS Lett. 2006, 580, 2811–2820. [Google Scholar] [CrossRef] [PubMed]
- Cox, T.R.; Erler, J.T. Remodeling and homeostasis of the extracellular matrix: Implications for fibrotic diseases and cancer Disease. Models Mech. 2011, 4, 165–178. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, S.; Takahashi, K. Induction of pluripotent stem cells from mouse fibroblast cultures. Tanpakushitsu Kakusan Koso 2006, 51, 2346–2351. [Google Scholar] [PubMed]
- Yu, J.; Vodyanik, M.A.; Smuga-Otto, K.; Antosiewicz-Bourget, J.; Frane, J.L.; Tian, S.; Nie, J.; Jonsdottir, G.A.; Ruotti, V.; Stewart, R.; et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 2007, 318, 1917–1920. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanley, J.; Rastegarlari, G.; Nathwani, A.C. An introduction to induced pluripotent stem cells. Br. J. Haematol. 2010, 151, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Anokye-Danso, F.; Snitow, M.; Morrisey, E.E. How microRNAs facilitate reprogramming to pluripotency. J. Cell Sci. 2012, 125, 4179–4787. [Google Scholar] [CrossRef] [PubMed]
- Razak, S.R.; Ueno, K.; Takayama, N.; Nariai, N.; Nagasaki, M.; Saito, R.; Koso, H.; Lai, C.; Murakami, M.; Tsuji, K.; et al. Profiling of microRNA in human and mouse ES and iPS cells reveals overlapping but distinct microRNA expression patterns. PLoS ONE 2013, 8, 73532. [Google Scholar] [CrossRef] [PubMed]
- Anokye-Danso, F.; Trivedi, C.M.; Juhr, D.; Gupta, M.; Cui, Z.; Tian, Y.; Zhang, Y.; Yang, W.; Gruber, P.J.; Epstein, J.A.; et al. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell 2011, 8, 376–388. [Google Scholar] [CrossRef] [PubMed]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef] [PubMed]
- Saint-André, V.; Federation, A.J. Models of human core transcriptional regulatory circuitries. Genome Res. 2015. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Duan, S.; Yi, F.; Yi, F.; Ocampo, A.; Liu, L.; Belmonte, J.C. Mitochondrial Regulation in Pluripotent Stem Cells. Cell Metab. 2013, 18, 325–332. [Google Scholar] [CrossRef] [PubMed]
- Kaspi, H.; Chapnik, E.; Levy, M.; Beck, G.; Hornstein, E.; Soen, Y. Brief Report: miR-290–295 Regulate Embryonic Stem Cell Differentiation Propensities by Repressing Pax6. Stem Cells 2013, 31, 2266–2272. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Blelloch, R. Cell Cycle Regulation by MicroRNAs in Embryonic Stem Cells. Cancer Res. 2009, 15, 4093–4096. [Google Scholar] [CrossRef] [PubMed]
- Gambardella, G.; Carissimo, A.; Chen, A.; Cutillo, L.; Nowakowski, T.J.; di Bernardo, D.; Blelloch, R. The impact of microRNAs on transcriptional heterogeneity and gene co-expression across single embryonic stem cells. Nat. Commun. 2017, 8, 14126. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.M.; Cahan, P.; Shalek, A.K.; Satija, R.; DaleyKeyser, A.J.; Li, H.; Zhang, J.; Pardee, K.; Gennert, D.; Trombetta, J.J.; et al. Deconstructing transcriptional heterogeneity in pluripotent stem cells. Nature 2014, 516, 56–61. [Google Scholar] [CrossRef] [PubMed]
- DeCastro, A.J.; DiRenzo, J. miRNA function and modulation in stem cells and cancer stem cells. microRNA Diagn. Ther. 2014, 12–17. [Google Scholar] [CrossRef]
- Ambros, V.; Chen, X. The regulation of genes and genomes by small RNAs. Development 2007, 134, 1635–1641. [Google Scholar] [CrossRef] [PubMed]
- Medeiros, L.A.; Dennis, L.M.; Gill, M.E.; Houbaviy, H.; Markoulaki, S.; Fu, D.; White, A.C.; Kirak, O.; Sharp, P.A.; Page, D.C.; et al. Mir-290-295 deficiency in mice results in partially penetrant embryonic lethality and germ cell defects. Proc. Natl. Acad. Sci. USA 2011, 108, 14163–14168. [Google Scholar] [CrossRef] [PubMed]
- Suh, M.R.; Lee, Y.; Kim, J.Y.; Kim, S.K.; Moon, S.H.; Lee, J.Y.; Cha, K.Y.; Chung, H.M.; Yoon, H.S.; Moon, S.Y.; et al. Human embryonic stem cells express a unique set of microRNAs. Dev. Biol. 2004, 270, 488–498. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.L.; Chang, D.C.; Chang-Lin, S.; Lin, C.H.; Wu, D.T.; Chen, D.T.; Ying, S.Y. Mir-302 reprograms human skin cancer cells into a pluripotent ES-cell-like state. RNA 2008, 14, 2115–2124. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, N.; Ishii, H.; Nagano, H.; Haraguchi, N.; Dewi, D.L.; Kano, Y.; Nishikawa, S.; Tanemura, M.; Mimori, K.; Tanaka, F.; et al. Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell 2011, 8, 633–638. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.J.; Lin, C.P.; Ho, J.J.; He, X.; Okada, N.; Bu, P.; Zhong, Y.; Kim, S.Y.; Bennett, M.J.; Chen, C.; et al. miR-34 miRNAs provide a barrier for somatic cell reprogramming. Nat. Cell Biol. 2011, 13, 1353–1360. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, Z.; Jiang, J.; Xu, C.; Kang, J.; Xiao, L.; Wu, M.; Xiong, J.; Guo, X.; Liu, H. Endogenous miRNA Sponge lincRNA-RoR Regulates Oct4, Nanog, and Sox2 in Human Embryonic Stem Cell Self-Renewal. Dev. Cell 2013, 25, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.K.; Kagalwala, M.N.; Parker-Thornburg, J.; Adams, H.; Majumder, S. REST maintains selfrenewal and pluripotency of embryonic stem cells. Nature 2008, 453, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Liang, J.; Ni, S.; Zhou, T.; Qing, X.; Li, H.; He, W.; Chen, J.; Li, F.; Zhuang, Q.; et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell 2010, 7, 51–63. [Google Scholar] [CrossRef] [PubMed]
- Korpal, M.; Kang, Y. The emerging role of miR-200 family of microRNAs in epithelial-mesenchymal transition and cancer metastasis. RNA Biol. 2008, 5, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Samavarchi-Tehrani, P.; Golipour, A.; David, L.; Sung, H.K.; Beyer, T.A.; Datti, A.; Woltjen, K.; Nagy, A.; Wrana, J.L. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell 2010, 7, 64–77. [Google Scholar] [CrossRef] [PubMed]
- Mani, S.A.; Guo, W.; Liao, M.J.; Eaton, E.N.; Ayyanan, A.; Zhou, A.Y.; Brooks, M.; Reinhard, F.; Zhang, C.C.; Shipitsin, M.; et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008, 133, 704–715. [Google Scholar] [CrossRef] [PubMed]
- Dumont, N.; Wilson, M.B.; Crawford, Y.G.; Reynolds, P.A.; Sigaroudinia, M.; Tlsty, T.D. Sustained induction of epithelial to mesenchymal transition activates DNA methylation of genes silenced in basal-like breast cancers. Proc. Natl. Acad. Sci. USA 2008, 105, 14867–14872. [Google Scholar] [CrossRef] [PubMed]
- Vrba, L.; Jensen, T.J.; Garbe, J.C.; Heimark, R.L.; Cress, A.E.; Dickinson, S.; Stampfer, M.R.; Futscher, B.W. Role for DNA methylation in the regulation of miR-200c and miR-141 expression in normal and cancer cells. PLoS ONE 2010, 5, e8697. [Google Scholar] [CrossRef] [PubMed]
- Wiklund, E.D.; Gao, S.; Hulf, T.; Sibbritt, T.; Nair, S.; Costea, D.E.; Villadsen, S.B.; Bakholdt, V.; Bramsen, J.B.; Sørensen, J.A.; et al. MicroRNA alterations and associated aberrant DNA methylation patterns across multiple sample types in oral squamous cell carcinoma. PLoS ONE 2011, 6, e27840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiklund, E.D.; Bramsen, J.B.; Hulf, T.; Dyrskjøt, L.; Ramanathan, R.; Hansen, T.B.; Villadsen, S.B.; Gao, S.; Ostenfeld, M.S.; Borre, M.; et al. Coordinated epigenetic repression of the miR-200 family and miR-205 in invasive bladder cancer. Int. J. Cancer 2011, 128, 1327–1334. [Google Scholar] [CrossRef] [PubMed]
- Gregory, P.A.; Bracken, C.P.; Smith, E.; Bert, A.G.; Wright, J.A.; Roslan, S.; Morris, M.; Wyatt, L.; Farshid, G.; Lim, Y.Y.; et al. An autocrine TGF-beta/ZEB/miR-200 signaling network regulates establishment and maintenance of epithelial-mesenchymal transition. Mol. Biol. Cell. 2011, 15, 1686–1698. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Han, Q.; Pei, D. EMT and MET as paradigms for cell fate switching. J. Mol Cell Biol. 2012, 4, 66–69. [Google Scholar] [CrossRef] [PubMed]
- Iliopoulos, D.; Lindahl-Allen, M.; Polytarchou, C.; Hirsch, H.A.; Tsichlis, P.N.; Struhl, K. Loss of miR-200 inhibition of Suz12 leads to polycomb-mediated repression required for the formation and maintenance of cancer stem cells. Mol. Cell 2010, 39, 761–772. [Google Scholar] [CrossRef] [PubMed]
- An, J.; Zheng, Y.; Tenenhaus Dann, C. Mesenchymal to Epithelial Transition Mediated by CDH1 Promotes Spontaneous Reprogramming of Male Germline Stem Cells to Pluripotency. Stem Cell Rep. 2017, 8, 446–459. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Kim, N.; Park, S.-W.; Kim, H.; Park, H.-J.; Han, Y.-M. Lineage-specific Expression of miR-200 Family in Human Embryonic Stem Cells during In Vitro Differentiation. Int. J. Stem Cells 2017, 10, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Eguchi, T.; Kuboki, T. Cellular Reprogramming Using Defined Factors and MicroRNAs. Stem Cells Int. 2016. [Google Scholar] [CrossRef] [PubMed]
- Hill, L.; Browne, G.; Tulchinsky, E. ZEB/miR-200 feedback loop: At the crossroads of signal transduction in cancer. Int. J. Cancer. 2013, 132, 745–754. [Google Scholar] [CrossRef] [PubMed]
- Subramanyam, D.; Lamouille, S.; Judson, R.L.; Liu, J.Y.; Bucay, N.; Derynck, R.; Blelloch, R. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat. Biotechnol. 2011, 29, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Zhu, X.; Dou, Y. The miR-302/367 cluster: A comprehensive update on its evolution and functions. Open Biol. 2015, 5, 150138. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Aksoy, M.; Shi, J.; Houbaviy, H. Evolution of the miR-/miR-371–373 Cluster Family Seed Repertoire. PLoS ONE 2014, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Roybon, L.; Lamas, N.J.; Garcia, A.D.; Yang, E.J.; Sattler, R.; Lewis, V.J.; Kim, Y.A.; Kachel, C.A.; Rothstein, J.D.; Przedborski, S.; et al. Human stem cell-derived spinal cord astrocytes with defined mature or reactive phenotypes. Cell Rep. 2013, 4, 1035–1048. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Baskerville, S.; Shenoy, A.; Babiarz, J.E.; Baehner, L.; Blelloch, R. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat. Genet. 2008, 40, 1478–1483. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.X.; Ravi, A.; Calabrese, J.M.; Medeiros, L.A.; Kirak, O.; Dennis, L.M.; Jaenisch, R.; Burge, C.B.; Sharp, P.A. A latent pro-survival function for the mir-290–295 cluster in mouse embryonic stem cells. PLoS Genet. 2011, 7, e1002054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jesus, A.B.D.; Lucena-Aguilar, G.; Menendez, P. The miR-302–367 cluster as a potential stemness regulator in ESCs. Cell Cycle 2009, 8, 394–398. [Google Scholar] [CrossRef] [PubMed]
- Card, D.A.; Hebbar, P.B.; Li, L.; Trotter, K.W.; Komatsu, Y.; Mishina, Y.; Archer, T.K. Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Mol. Cell. Biol. 2008, 28, 6426–6438. [Google Scholar] [CrossRef] [PubMed]
- Lange, C.; Huttner, W.B.; Calegari, F. Cdk4/cyclinD1 overexpression in neural stem cells shortens G1, delays neurogenesis, and promotes the generation and expansion of basal progenitors. Cell Stem Cell 2009, 5, 320–331. [Google Scholar] [CrossRef] [PubMed]
- Kuo, Y.C.; Weng, S.C.; Chou, C.J.; Chang, T.T.; Tsai, W.J. Activation and proliferation signals in primary human T lymphocytes inhibited by ergosterol peroxide isolated from Cordyceps cicadae. Br. J. Pharmacol. 2003, 140, 895–906. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.L.; Chang, D.C.; Lin, C.H.; Ying, S.Y.; Leu, D.; Wu, D.T.S. Regulation of somatic cell reprogramming through inducible mir-302 expression. Nucleic Acids Res. 2011, 39, 1054–1065. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.R.; Prasain, N.; Chae, H.D.; Kim, Y.J.; Mantel, C.; Yoder, M.C.; Broxmeyer, H.E. Epigenetic regulation of NANOG by miR-302 cluster-MBD2 completes induced pluripotent stem cell reprogramming. Stem Cells 2013, 31, 666–681. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, J.W.; Gomez, E.W. Biomechanics of TGFbeta-induced epithelial–mesenchymal transition: Implications for fibrosis and cancer. Clin. Transl. Med. 2014, 3, 23. [Google Scholar] [CrossRef] [PubMed]
- Barroso-delJesus, A.; Lucena-Aguilar, G.; Sanchez, L.; Ligero, G.; Gutierrez-Aranda, I.; Menendez, P. The Nodal inhibitor Lefty is negatively modulated by the microRNA miR-302 in human embryonic stem cells. FASEB J. 2011, 25, 1497–1508. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Cui, W. Sox2, a key factor in the regulation of pluripotency and neural differentiation. World J. Stem Cells 2014, 6, 305–311. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-S.; Yi, B.-R.; Kim, N.-H.; Choi, K.-C. Role of the epithelial–mesenchymal transition and its effects on embryonic stem cells. Exp. Mol. Med. 2014, 46, e108. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Hutchins, A.P.; Chen, Y.; Li, S.; Shan, Y.; Liao, B.; Zheng, D.; Shi, X.; Li, Y.; Chan, W.-Y.; et al. A sequential EMT-MET mechanism drives the differentiation of human embryonic stem cells towards hepatocytes. Nat. Commun. 2017, 8, 15166. [Google Scholar] [CrossRef] [PubMed]
- Ichida, J.K.; Blanchard, J.; Lam, K.; Son, E.Y.; Chung, J.E.; Egli, D.; Loh, K.M.; Carter, A.C.; di Giorgio, F.P.; Koszka, K.; et al. A small-molecule inhibitor of TGF-Beta signaling replaces Sox2 in reprogramming by inducing nanog. Cell Stem Cell 2009, 5, 491–503. [Google Scholar] [CrossRef] [PubMed]
- Young, J.L.; Suresh, R.; Chauhan, H.; Park, W.S.; Hong, S.-H.; Kim, K.-S. Dissecting microRNA-mediated regulation of stemness, reprogramming, and pluripotency. Cell Regen. 2016, 5, 2. [Google Scholar]
- Chan, Y.C.; Khanna, S.; Roy, S.; Sen, C.K. miR-200b Targets Ets-1 and Is Down-regulated by Hypoxia to Induce Angiogenic Response of Endothelial Cells. J. Biol. Chem. 2011, 286, 2047–2056. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.-C.; Yeh, C.-H. Inhibition of miR-302 Suppresses Hypoxia-Reoxygenation-Induced H9c2 Cardiomyocyte Death by Regulating Mcl-1 Expression. Oxid. Med. Cell. Longev. 2017. [Google Scholar] [CrossRef] [PubMed]
- Ji, A.R.; Ku, S.Y.; Cho, M.S.; Kim, Y.Y.; Kim, Y.J.; Oh, S.K.; Kim, S.H.; Moon, S.Y.; Choi, Y.M. Reactive oxygen species enhance differentiation of human embryonic stem cells into mesendodermal lineage. Exp. Mol. Med. 2010, 42, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Maioli, M.; Rinaldi, S.; Santaniello, S.; Castagna, A.; Pigliaru, G.; Gualini, S.; Cavallini, C.; Fontani, V.; Ventura, C. Radio Electric Conveyed Fields Directly Reprogram Human Dermal Skin Fibroblasts Toward Cardiac, Neuronal, and Skeletal Muscle-Like Lineages. Cell Transplant. 2013, 22, 1227–1235. [Google Scholar] [CrossRef] [PubMed]
- Maioli, M.; Rinaldi, S.; Pigliaru, G.; Santaniello, S.; Basoli, V.; Castagna, A.; Fontani, V.; Ventura, C. REAC technology and hyaluron synthase 2, an interesting network to slow down stem cell senescence. Sci. Rep. 2016, 6, 28682. [Google Scholar] [CrossRef] [PubMed]
- Maioli, M.; Rinaldi, S.; Santaniello, S.; Castagna, A.; Pigliaru, G.; Delitala, A.; Bianchi, F.; Tremolada, C.; Fontani, V.; Ventura, C. Radioelectric asymmetric conveyed fields and human adipose-derived stem cells obtained with a nonenzymatic method and device: A novel approach to multipotency. Cell Transplant. 2014, 23, 1489–1500. [Google Scholar] [CrossRef] [PubMed]
- Baek, S.; Quan, X.; Kim, S.; Lengner, C.; Park, J.K.; Kim, J. Electromagnetic fields mediate efficient cell reprogramming into a pluripotent state. ACS Nano 2014, 8, 10125–10138. [Google Scholar] [CrossRef] [PubMed]
- Razavi, S.; Salimi, M.; Shahbazi-Gahrouei, D.; Karbasi, S.; Kermani, S. Extremely low-frequency electromagnetic field influences the survival and proliferation effect of human adipose derived stem cells. Adv. Biomed. Res. 2014, 3, 25. [Google Scholar] [CrossRef] [PubMed]
- Baek, S.; Oh, J.; Song, J.; Choi, H.; Yoo, J.; Park, G.Y.; Han, J.; Chang, Y.; Park, H.; Kim, H.; et al. Generation of Integration-Free Induced Neurons Using Graphene Oxide-Polyethylenimine. Small 2017, 13, 5. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Wu, W.; Chen, X.; He, C.; Liu, X. Effect of Pulsed Electromagnetic Field with Different Frequencies on the Proliferation, Apoptosis and Migration of Human Ovarian Cancer Cells. J. Biomed. Eng. 2012, 29, 291–295. [Google Scholar]
- Juutilainen, J. Developmental Effects of Electromagnetic Fields. Bioelectromagnetics 2005, 26, S107–S115. [Google Scholar] [CrossRef] [PubMed]
- Levin, M. Large-Scale Biophysics: Ion Flows and Regeneration. Trends Cell Biol. 2007, 17, 261–270. [Google Scholar] [CrossRef] [PubMed]
- Hiraki, Y.; Endo, N.; Takigawa, M.; Asada, A.; Takahashi, H.; Suzuki, F. Enhanced Responsiveness to Parathyroid Hormoneand Induction of Functional Differentiation of Cultured Rabbit Costal Chondrocytes by a Pulsed Electromagnetic Field. Biochim. Biophys. Acta 1987, 931, 94–100. [Google Scholar] [CrossRef]
- Kang, K.S.; Hong, J.M.; Kang, J.A.; Rhie, J.W.; Jeong, Y.H.; Cho, D.W. Regulation of Osteogenic Differentiation of Human Adipose-Derived Stem Cells by Controlling Electromagnetic Field Conditions. Exp. Mol. Med. 2013, 45, e6. [Google Scholar] [CrossRef] [PubMed]
- Meng, D.; Xu, T.; Guo, F.; Yin, W.; Peng, T. The Effects ofHigh-Intensity Pulsed Electromagnetic Field on Proliferationand Differentiation of Neural Stem Cells of Neonatal Rats in vitro. Med. Sci. 2009, 29, 732–736. [Google Scholar]
- Sun, L.Y.; Hsieh, D.K.; Lin, P.C.; Chiu, H.T.; Chiou, T.W. Pulsed electromagnetic fields accelerate proliferation and osteogenic gene expression in human bone marrow mesenchymal stem cells during osteogenic differentiation. Bioelectromagnetics 2010, 31, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, Z.; Fisher, M.; Lohmann, C.H.; Simon, B.J.; Boya, B.D. Osteoprotegerin (OPG) production by cells in the osteoblast lineage is regulated by pulsed electromagnetic fields in cultures grown on calcium phosphate substrates. Ann. Biomed. Eng. 2009, 37, 437–444. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, Z.; Simon, B.J.; Duran, M.A.; Barabino, G.; Chaudhri, R.; Boyan, B.D. Pulsed electromagnetic fields enhance BMP-2 dependent osteoblastic differentiation of human mesenchymal stem cells. J. Orthop. Res. 2008, 26, 1250–1255. [Google Scholar] [CrossRef] [PubMed]
- Janssen-Heininger, Y.M.; Mossman, B.T.; Heintz, N.H.; Forman, H.J.; Kalyanaraman, B.; Finkel, T.; Stamler, J.S.; Rhee, S.G.; van der Vliet, A. Redox-based regulation of signal transduction: Principles, pitfalls, and promises. Free Radic. Biol. Med. 2008, 45, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Maziarz, A.; Kocan, B.; Bester, M.; Budzik, S.; Cholewa, M.; Ochiya, T.; Banas, A. How electromagnetic fields can influence adult stem cells: Positive and negative impacts. Stem Cell Res. Ther. 2016, 7, 54. [Google Scholar] [CrossRef] [PubMed]
- Holmström, K.M.; Finkel, T. Cellular mechanisms and physiological consequences of redox-dependent signaling. Nat. Rev. Mol. Cell Biol. 2014, 15, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Nagai, H.; Lin, T.M.; Peterson, R.E.; Tohyama, C.; Kobayashi, T.; Nohara, K. Organic Chemicals Adsorbed onto Diesel Exhaust Particles Directly Alter the Differentiation of Fetal Thymocytes Through Arylhydrocarbon Receptor but Not Oxidative Stress Responses. J. Immunotoxicol. 2006, 3, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Milhavet, O.; Lemaitre, J.M. Senescent-Derived Pluripotent Stem Cells Are Able to Redifferentiate into Fully Rejuvenated Cells. In Tumor Dormancy, Quiescence, and Senescence; Springer: Dordrecht, The Netherlands, 2014; Volume 2, pp. 265–276. [Google Scholar]
- Teslaa, T.; Teitell, M.A. Pluripotent stem cell energy metabolism: An update. EMBO J. 2015, 34, 138–153. [Google Scholar] [CrossRef] [PubMed]
- Ezashi, T.; Das, P.; Roberts, R.M. Low O2 tensions andthe prevention of differentiation of hES cells. Proc. Natl. Acad. Sci. USA 2005, 102, 4783–4788. [Google Scholar] [CrossRef] [PubMed]
- Westfall, S.D.; Sachdev, S.; Das, P.; Hearne, L.B.; Hannink, M.; Roberts, R.M.; Ezashi, T. Identification of Oxygen-Sensitive Transcriptional Programs in Human Embryonic Stem Cells. Stem Cells Dev. 2008, 17, 869–881. [Google Scholar] [CrossRef] [PubMed]
- Dando, I.; Cordani, M.; DallaPozza, E.; Biondani, G.; Donadelli, M.; Palmieri, M. Antioxidant Mechanisms and ROS-Related MicroRNAs in Cancer Stem CellsOxid. Med. Cell Longev. 2015, 425708. [Google Scholar]
- Vaupel, P.; Mayer, A. Hypoxia in cancer: Significance and impact on clinical outcome. Cancer Metastasis Rev. 2007, 26, 225–239. [Google Scholar] [CrossRef] [PubMed]
- Höckel, M.; Vaupel, P. Tumor hypoxia: Definitions and current clinical, biologic, and molecular aspects. J. Natl. Cancer Inst. 2001, 93, 266–276. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.-M.; Desai, L.P. Reciprocal regulation of TGF-β and reactive oxygen species: A perverse cycle for fibrosis. Redox Biol. 2015, 6, 565–577. [Google Scholar] [CrossRef] [PubMed]
- Magenta, A.; Cencioni, C.; Fasanaro, P.; Zaccagnini, G.; Greco, S.; Sarra-Ferraris, G.; Antonini, A.; Martelli, F.; Capogrossi, M.C. miR-200c is upregulated by oxidative stress and induces endothelial cell apoptosis and senescence via ZEB1 inhibition. Cell Death Differ. 2011, 18, 1628–1639. [Google Scholar] [CrossRef] [PubMed]
- Ross, C.L.; Siriwardane, M.; Almeida-Poradaa, G.; Poradaa, C.D.; Brinkc, P.; Christa, G.J.; Harrisona, B.S. The effect of low-frequency electromagnetic field on human bone marrow stem/progenitor cell differentiation. Stem Cell Res. 2015, 15, 96–108. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, E.J.; Baldwin, L.A. Defining hormesis. Hum. Exp. Toxicol. 2002, 21, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Shin, K.K.; Lee, A.L.; Kim, Y.S.; Park, H.J.; Park, Y.K.; Bae, Y.C.; Jung, J.S. MicroRNA-302 induces proliferation and inhibits oxidant-induced cell death in human adipose tissue-derived mesenchymal stem cells. Cell Death Dis. 2014, 5, e1385. [Google Scholar] [CrossRef] [PubMed]
- Harnoss, J.M.; Strowitzki, M.J.; Radhakrishnan, P.; Platzer, L.K.; Harnoss, J.C.; Hank, T.; Cai, J.; Ulrich, A.; Schneider, M. Therapeutic inhibition of prolyl hydroxylase domain-containing enzymes in surgery: Putative applications and challenges. Hypoxia 2015, 3, 1–14. [Google Scholar] [PubMed]

| miRNA | Influence on Pluripontecy | Influence on Differentiation | Inhibition of EMT /MET |
|---|---|---|---|
| miR-200 family | yes | no | yes |
| miR-302 family | yes | no | yes |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Balzano, F.; Cruciani, S.; Basoli, V.; Santaniello, S.; Facchin, F.; Ventura, C.; Maioli, M. MiR200 and miR302: Two Big Families Influencing Stem Cell Behavior. Molecules 2018, 23, 282. https://doi.org/10.3390/molecules23020282
Balzano F, Cruciani S, Basoli V, Santaniello S, Facchin F, Ventura C, Maioli M. MiR200 and miR302: Two Big Families Influencing Stem Cell Behavior. Molecules. 2018; 23(2):282. https://doi.org/10.3390/molecules23020282
Chicago/Turabian StyleBalzano, Francesca, Sara Cruciani, Valentina Basoli, Sara Santaniello, Federica Facchin, Carlo Ventura, and Margherita Maioli. 2018. "MiR200 and miR302: Two Big Families Influencing Stem Cell Behavior" Molecules 23, no. 2: 282. https://doi.org/10.3390/molecules23020282






