Synthesis, DNA Binding, and Anticancer Properties of Bis-Naphthalimide Derivatives with Lysine-Modified Polyamine Linkers
Abstract
:1. Introduction
2. Results and Discussion
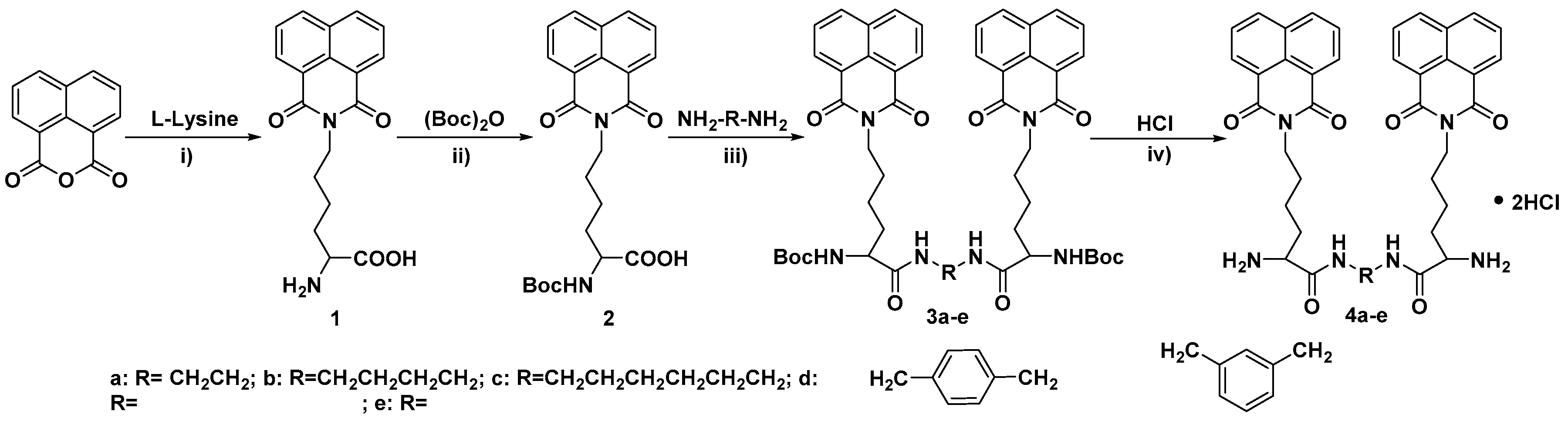
2.1. Chemistry
2.2. UV–Vis Titration
2.3. DNA Binding Selectivity
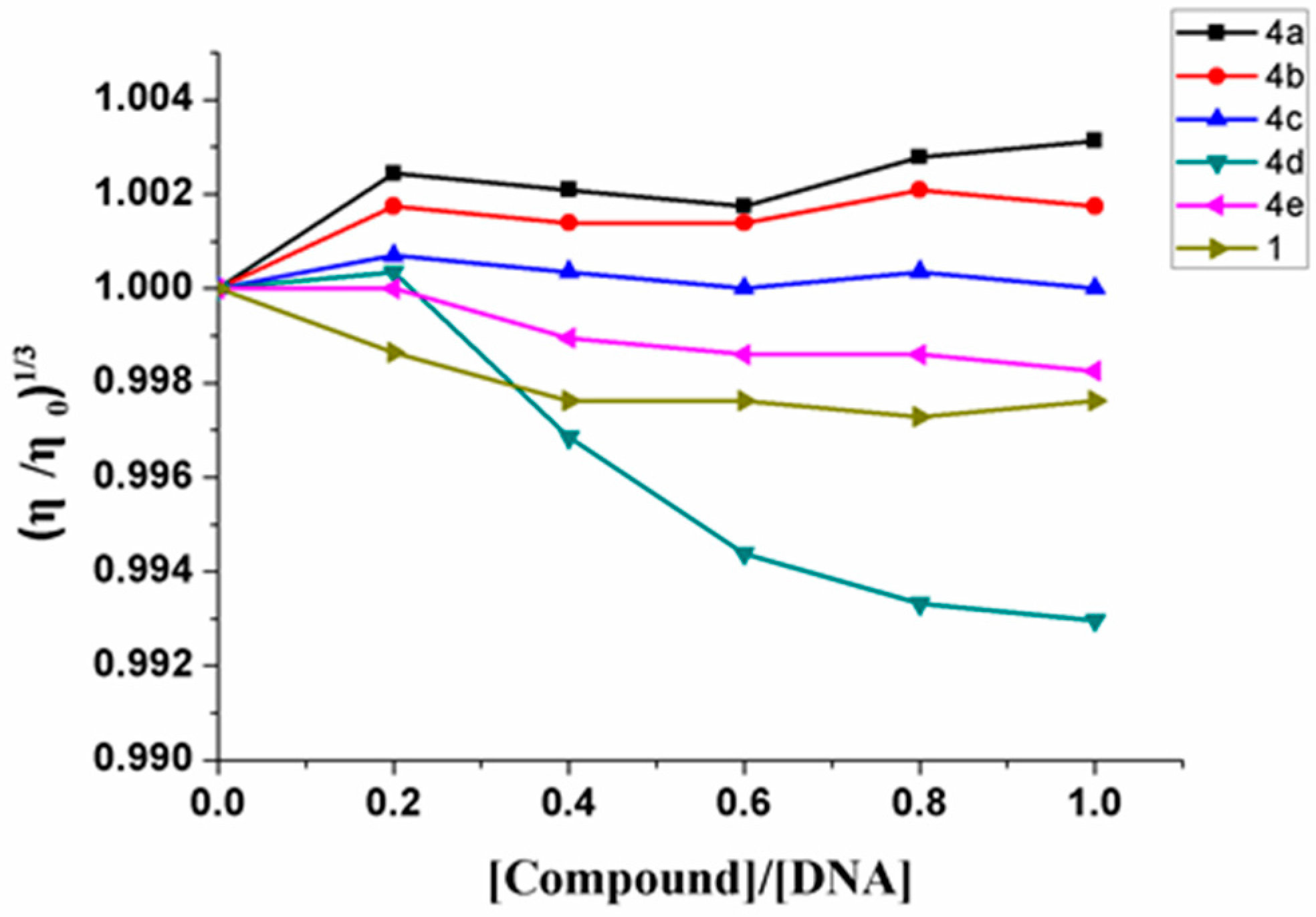
2.4. Viscosity Studies
2.5. Cytotoxicity Assay
2.6. Morphology Observation
3. Materials and Methods
3.1. Chemistry
3.1.1. Synthesis of N-Epsilon-1,8-naphthalimido-lysine (1)
3.1.2. Synthesis of N-Alpha-(tert-Butoxycarbonyl)-N-epsilon-1,8-naphthalimido-lysine (2)
3.1.3. Synthesis of 3
3.1.4. Synthesis of 4
3.2. UV–Vis Titration
3.3. Viscosity Study
3.4. Cytotoxicity Assay
3.5. Morphology Observation
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Braña, M.F.; Ramos, A. Naphthalimides as anti-cancer agents: Synthesis and biological activity. Curr. Med. Chem. Anticancer Agents 2001, 1, 237–255. [Google Scholar] [CrossRef] [PubMed]
- Gellerman, G. Recent Developments in the Synthesis and Applications of Anticancer Amonafide Derivatives. A Mini Review. Lett. Drug Des. Discov. 2016, 13, 47–63. [Google Scholar] [CrossRef]
- Banerjee, S.; Veale, E.B.; Phelan, C.M.; Murphy, S.A.; Tocci, G.M.; Gillespie, L.J.; Frimannsson, D.O.; Kelly, J.M.; Gunnlaugsson, T. Recent advances in the development of 1,8-naphthalimide based DNA targeting binders, anticancer and fluorescent cellular imaging agents. Chem. Soc. Rev. 2013, 42, 1601–1618. [Google Scholar] [CrossRef] [PubMed]
- Norton, J.T.; Witschi, M.A.; Luong, L.; Kawamura, A.; Ghosh, S.; Stack, M.S.; Sim, E.; Avram, M.J.; Appella, D.H.; Huang, S. Synthesis and anticancer activities of 6-amino amonafide derivatives. Anticancer Drugs 2008, 19, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Lv, M.; Xu, H. Overview of naphthalimide analogs as anticancer agents. Curr. Med. Chem. 2009, 16, 4797–4813. [Google Scholar] [CrossRef] [PubMed]
- Freeman, C.L.; Swords, R.; Giles, F.J. Amonafide: A future in treatment of resistant and secondary acute myeloid leukemia? Expert Rev. Hematol. 2012, 5, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Stone, R.M.; Mazzola, E.; Neuberg, D.; Allen, S.L.; Pigneux, A.; Stuart, R.K.; Wetzler, M.; Rizzieri, D.; Erba, H.P.; Damon, L.; et al. Phase III open-label randomized study of cytarabine in combination with amonafide L-malate or daunorubicin as induction therapy for patients with secondary acute myeloid leukemia. J. Clin. Oncol. 2015, 33, 1252–1257. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.T.; Chen, T.L.; Chang, K.S.; Chang, C.M.; Wei, T.Y.; Liu, J.W.; Hsiao, C.A.; Shih, T.L. Synthesis of novel C4-benzazole naphthalimide derivatives with potent anti-tumor properties against murine melanoma. Bioorg. Med. Chem. 2017, 25, 789–794. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, J.; Li, M.; Li, M.; Xie, S.; Wang, C. Synthesis and evaluation of novel amonafide-polyamine conjugates as anticancer agents. Chem. Biol. Drug Des. 2017, 89, 670–680. [Google Scholar] [CrossRef] [PubMed]
- Ge, C.C.; Chang, L.P.; Zhao, Y.; Chang, C.C.; Xu, X.J.; He, H.Y.; Wang, Y.X.; Dai, F.J.; Xie, S.Q.; Wang, C.J. Design, Synthesis and Evaluation of Naphthalimide Derivatives as Potential Anticancer Agents for Hepatocellular Carcinoma. Molecules 2017, 22, 342. [Google Scholar] [CrossRef] [PubMed]
- Brider, T.; Redko, B.; Oron-Herman, M.; Cohen-Matzlich, A.; Gerlitz, G.; Gellerman, G.; Grynszpan, F. Synthesis and in vitro anticancer evaluation of 1,8-naphthalimide N(4) and S(4)-derivatives combining DNA intercalation and alkylation capabilities. Res. Chem. Intermed. 2016, 42, 1741–1757. [Google Scholar] [CrossRef]
- Bousquet, P.F.; Braña, M.F.; Conlon, D.; Fitzgerald, K.M.; Perron, D.; Cocchiaro, C.; Miller, R.; Moran, M.; George, J.; Qian, X.D.; et al. Preclinical evaluation of LU 79553: A novel bis-naphthalimide with potent antitumor activity. Cancer Res. 1995, 55, 1176–1180. [Google Scholar] [PubMed]
- Kamal, A.; Ramu, R.; Tekumalla, V.; Ramesh Khanna, G.B.; Zingde, S.M. Remarkable DNA binding affinity and potential anti-cancer activity of pyrrolo[2,1-c][1,4] benzodiazepine–naphthalimide conjugates linked through piperazine side-armed alkane spacers. Bioorg. Med. Chem. 2008, 16, 7218–7224. [Google Scholar] [CrossRef] [PubMed]
- Veale, E.B.; Frimannsson, D.O.; Lawler, M.; Gunnlaugsson, T. 4-Amino-1,8-naphthalimide-based Tröger’s bases as high affinity DNA targeting fluorescent supramolecular scaffolds. Org. Lett. 2009, 11, 4040–4043. [Google Scholar] [CrossRef] [PubMed]
- Veale, E.B.; Gunnlaugsson, T. Synthesis, photophysical, and DNA binding studies of fluorescent Tröger’s base derived 4-amino-1,8-naphthalimide supramolecular clefts. J. Org. Chem. 2010, 75, 5513–5525. [Google Scholar] [CrossRef] [PubMed]
- Antonini, I.; Santoni, G.; Lucciarini, R.; Amantini, C.; Sparapani, S.; Magnano, A. Synthesis and biological evaluation of new asymmetrical bisintercalators as potential antitumor drugs. J. Med. Chem. 2006, 49, 7198–7207. [Google Scholar] [CrossRef] [PubMed]
- Ott, I.; Xu, Y.; Qian, X. Fluorescence properties and antiproliferative effects of mono-, bis-, and tris-thiophenylnaphthalimides: Results of a comparative pilot study. J. Photochem. Photobiol. B 2011, 105, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Nagasawa, H.; Uto, Y.; Sugimoto, Y.; Noguchi, K.; Wakida, M.; Wierzba, K.; Terada, T.; Asao, T.; Yamada, Y.; et al. Napthalimidobenzamide DB-51630: A novel DNA binding agent inducing p300 gene expression and exerting a potent anti-cancer activity. Bioorg. Med. Chem. 2005, 13, 4014–4021. [Google Scholar] [CrossRef] [PubMed]
- Pavlov, V.; Kong Thoo Lin, P.; Rodilla, V. Cytotoxicity, DNA binding and localisation of novel bis-naphthalimidopropyl polyamine derivatives. Chem. Biol. Interact. 2001, 137, 15–24. [Google Scholar] [CrossRef]
- Lin, P.K.; Pavlov, V.A. The synthesis and in vitro cytotoxic studies of novel bis-naphthalimidopropyl polyamine derivatives. Bioorg. Med. Chem. Lett. 2000, 10, 1609–1612. [Google Scholar] [CrossRef]
- Barron, G.A.; Bermano, G.; Gordon, A.; Kong Thoo Lin, P. Synthesis, cytotoxicity and DNA-binding of novel bisnaphthalimidopropyl derivatives in breast cancer MDA-MB-231 cells. Eur. J. Med. Chem. 2010, 45, 1430–1437. [Google Scholar] [CrossRef] [PubMed]
- Filosa, R.; Peduto, A.; Micco, S.D.; Caprariis, P.; Festa, M.; Petrella, A.; Capranico, G.; Bifulco, G. Molecular modelling studies, synthesis and biological activity of a series of novel bis-naphthalimides and their development as new DNA topoisomerase II inhibitors. Bioorg. Med. Chem. 2009, 17, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Yang, Q.; Qian, X.B. Novel DNA bis-intercalators of isoquinolino[4,5-bc]acridines: Design, synthesis and evaluation of cytotoxic activity. Tetrahedron 2005, 61, 11895–11901. [Google Scholar] [CrossRef]
- Ralton, L.D.; Bestwick, C.S.; Milne, L.; Duthie, S.; Kong Thoo Lin, P. Bisnaphthalimidopropyl spermidine induces apoptosis within colon carcinoma cells. Chem. Biol. Interact. 2009, 177, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Rong, R.X.; Sun, Q.; Ma, C.L.; Chen, B.; Wang, W.Y.; Wang, Z.A.; Wang, K.R.; Cao, Z.R.; Li, X.L. Development of novel bis-naphthalimide derivatives and their anticancer properties. Med. Chem. Commun. 2016, 7, 679–685. [Google Scholar] [CrossRef]
- Wu, A.; Xu, Y.; Qian, X. Novel naphthalimide-amino acid conjugates with flexible leucine moiety as side chain: Design, synthesis and potential antitumor activity. Bioorg. Med. Chem. 2009, 17, 592–599. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Yang, P.; Qian, X.; Tong, L. Naphthalimide intercalators with chiral amino side chains: Effects of chirality on DNA binding, photodamage and antitumor cytotoxicity. Bioorg. Med. Chem. Lett. 2008, 18, 6210–6213. [Google Scholar] [CrossRef] [PubMed]
- McMasters, S.; Kelly, L.A. Sequence-dependent interactions of cationic naphthalimides and polynucleotides. Photochem. Photobiol. 2007, 83, 889–896. [Google Scholar] [CrossRef] [PubMed]
- Lerman, L.S. Structural considerations in the interaction of DNA and acridines. J. Mol. Biol. 1961, 3, 18–30. [Google Scholar] [CrossRef]
- Satyanarayana, S.; Dabrowiak, J.C.; Chaires, J.B. Tris(phenanthroline)ruthenium(II) enantiomer interactions with DNA: Mode and specificity of binding. Biochemistry 1993, 32, 2573–2584. [Google Scholar] [CrossRef] [PubMed]
- Kumar, C.V.; Asuncion, E.H. DNA Binding Studies and Sites Fluorescence Sensitization of an Anthryl Probe. J. Am. Chem. Soc. 1993, 115, 8547–8553. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds 1–4 are available from the authors. |

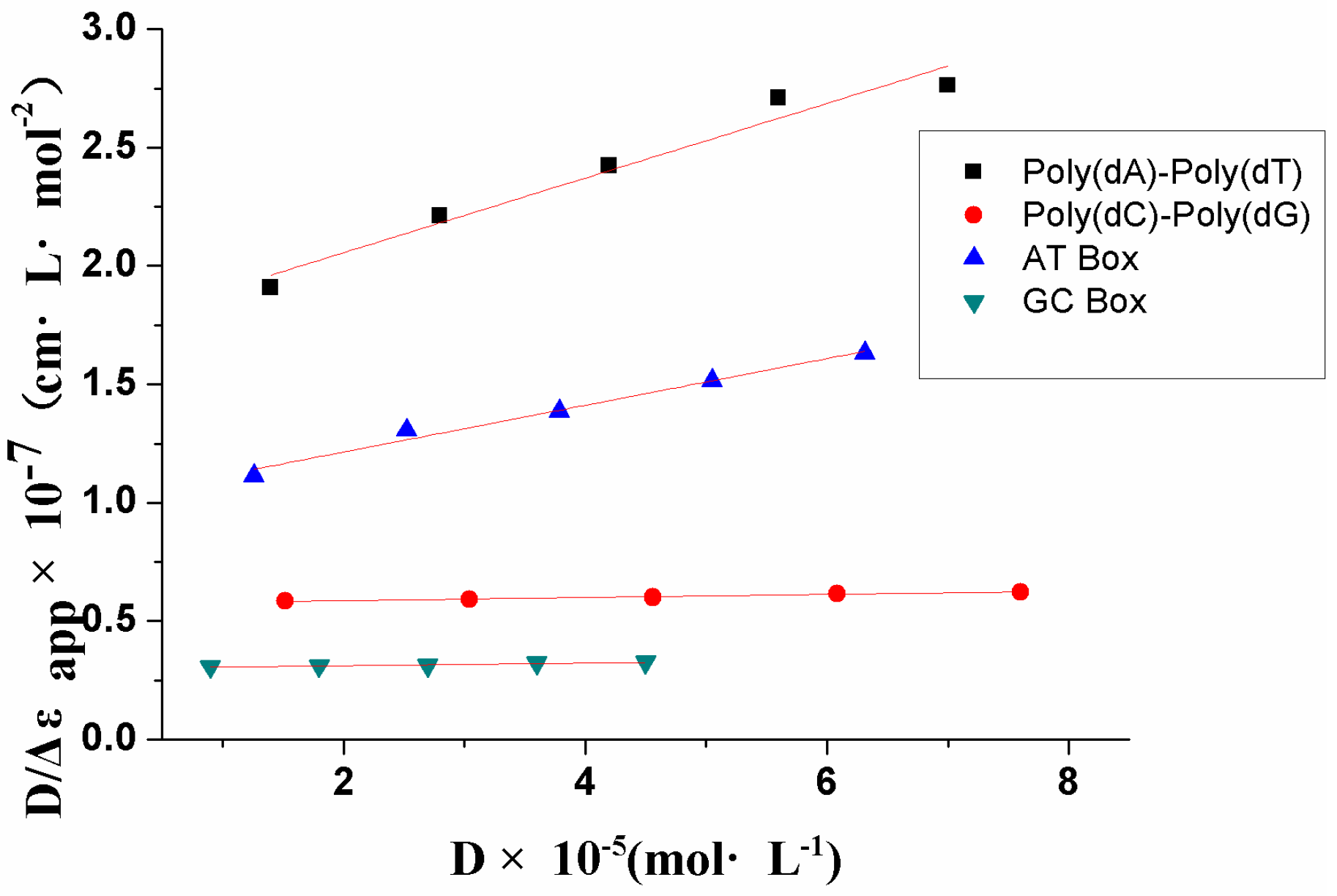

| Compound | Kb (M−1) |
|---|---|
| 4a | 3.40 × 104 |
| 4b | 2.78 × 104 |
| 4c | 1.54 × 104 |
| 4d | 7.29 × 103 |
| 4e | 1.00 × 104 |
| DNA Duplex | Kb (M−1) |
|---|---|
| [Poly(dA)-Poly(dT)] | 9.08 × 103 |
| [Poly(dG)-Poly(dC)] | 1.15 × 103 |
| AT Box | 9.71 × 103 |
| GC Box | 1.91 × 103 |
| Cell Lines | IC50 Value (μmol∙L−1) | |||||
|---|---|---|---|---|---|---|
| Fluorouracil | 4a | 4b | 4c | 4d | 4e | |
| EC109 | 45.82 | 694.59 | 456.35 | 330.38 | 142.45 | 354.48 |
| BGC823 | 49.88 | N.T. 1 | N.T. | 193.65 | 77.99 | N.T. |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Y.; Wu, C.-X.; Song, Y.; Huang, M.; Tian, D.-N.; Yang, X.-B.; Fan, Y.-R. Synthesis, DNA Binding, and Anticancer Properties of Bis-Naphthalimide Derivatives with Lysine-Modified Polyamine Linkers. Molecules 2018, 23, 266. https://doi.org/10.3390/molecules23020266
Huang Y, Wu C-X, Song Y, Huang M, Tian D-N, Yang X-B, Fan Y-R. Synthesis, DNA Binding, and Anticancer Properties of Bis-Naphthalimide Derivatives with Lysine-Modified Polyamine Linkers. Molecules. 2018; 23(2):266. https://doi.org/10.3390/molecules23020266
Chicago/Turabian StyleHuang, Yu, Chun-Xia Wu, Yu Song, Min Huang, Da-Nian Tian, Xin-Bin Yang, and Yan-Ru Fan. 2018. "Synthesis, DNA Binding, and Anticancer Properties of Bis-Naphthalimide Derivatives with Lysine-Modified Polyamine Linkers" Molecules 23, no. 2: 266. https://doi.org/10.3390/molecules23020266





