Complete Chloroplast Genome Sequence and Phylogenetic Analysis of Paeonia ostii
Abstract
:1. Introduction
2. Results and Discussion
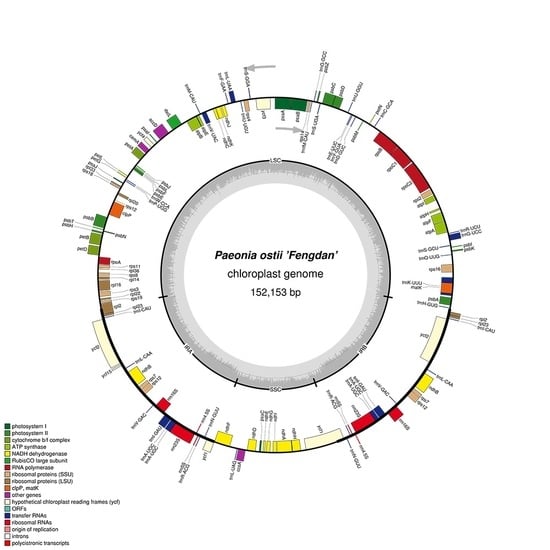
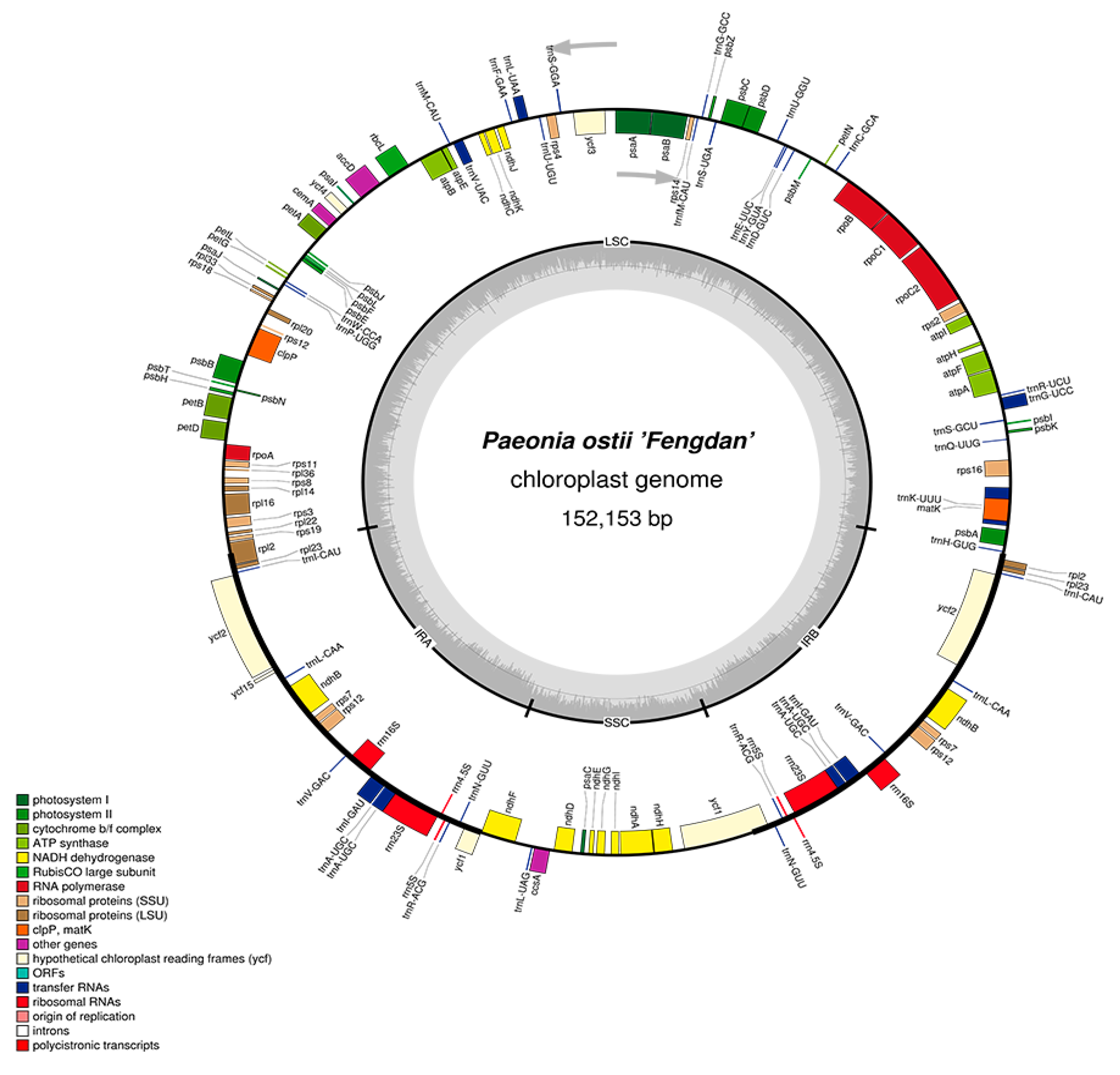
2.1. Features of P. ostii cpDNA
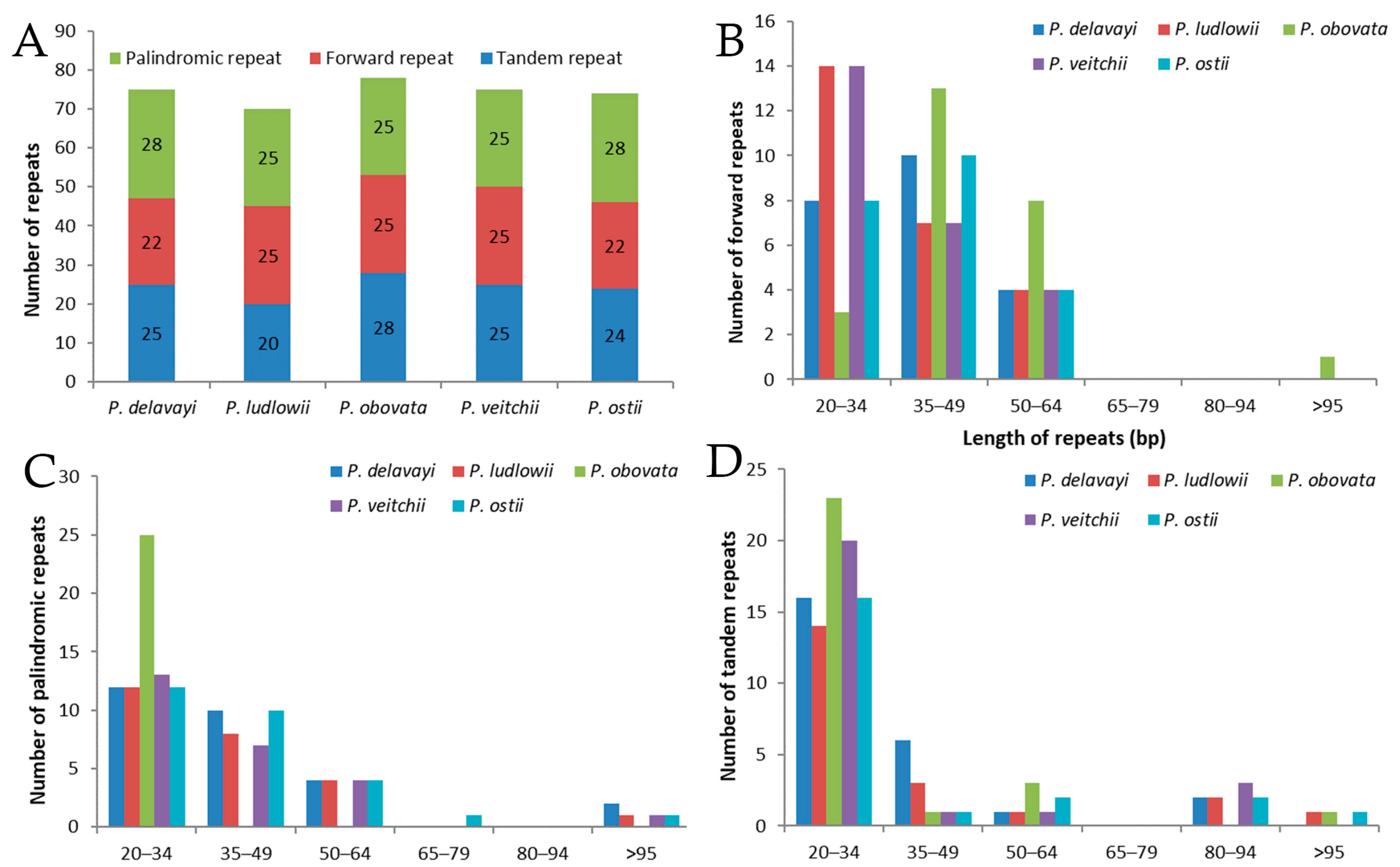
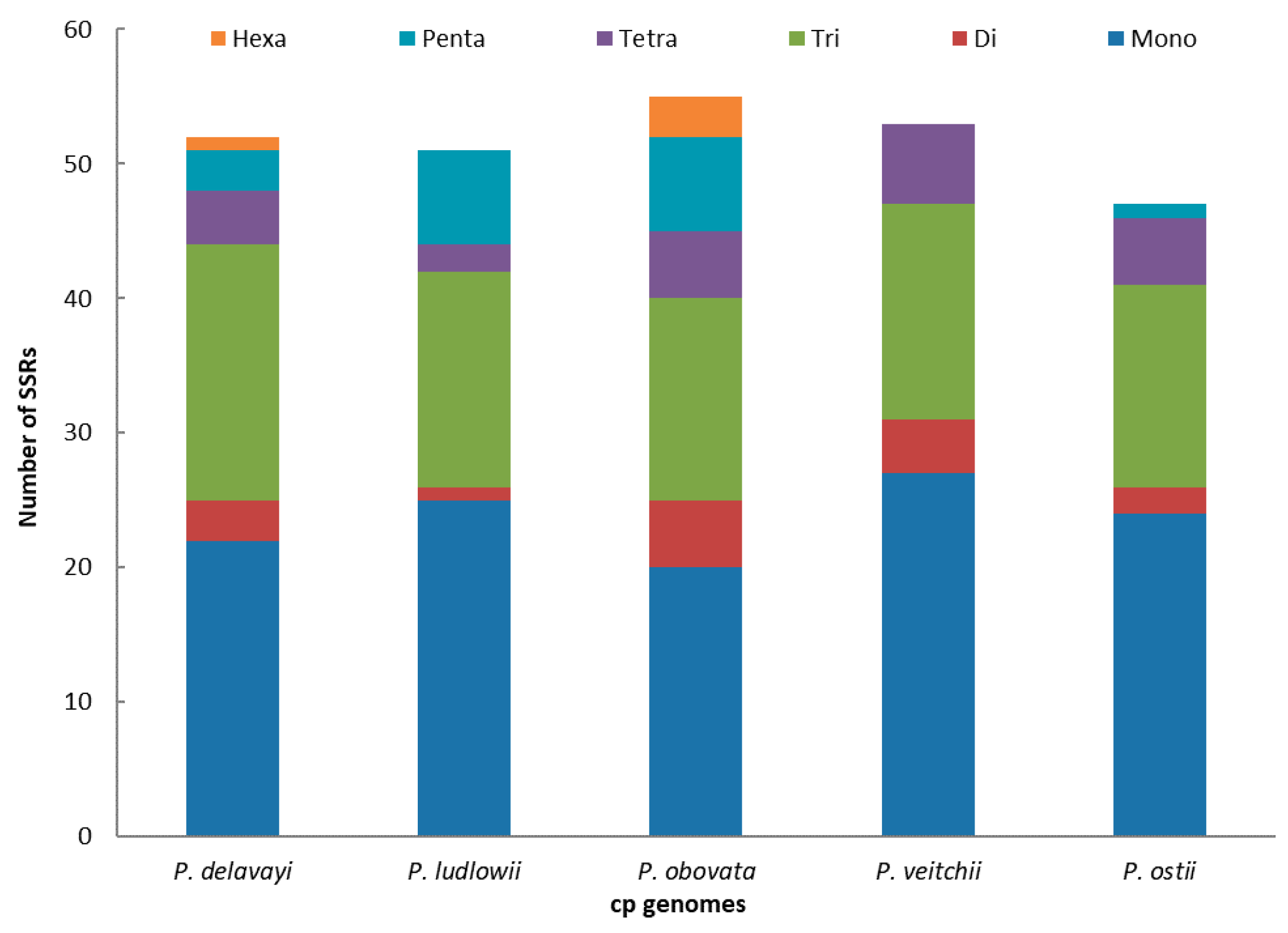
2.2. Long-Repeat and SSR Analysis
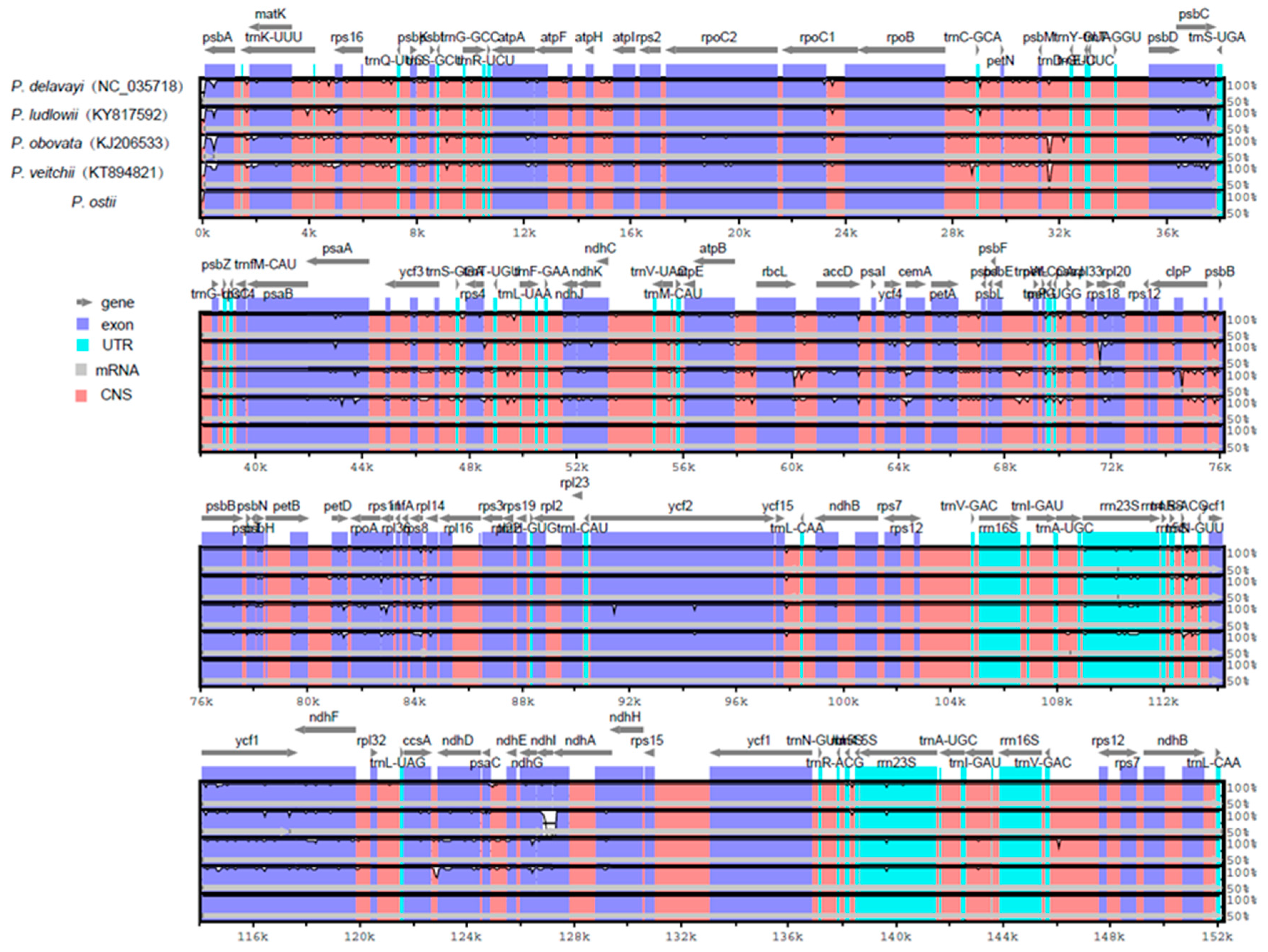
2.3. Comparative Chloroplast Genomic Analysis
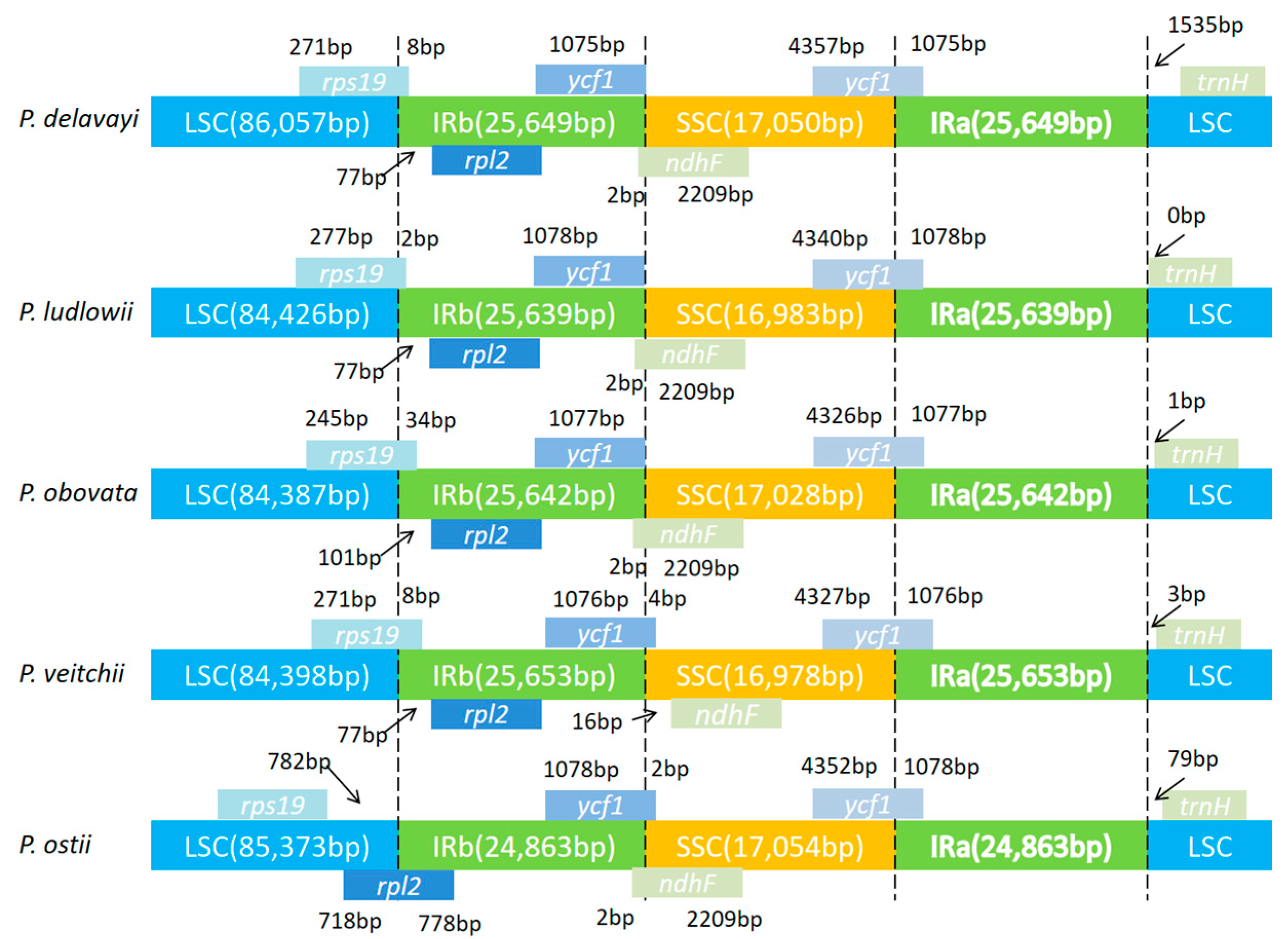
2.4. IR Contraction and Expansion in the P. ostii Chloroplast Genome
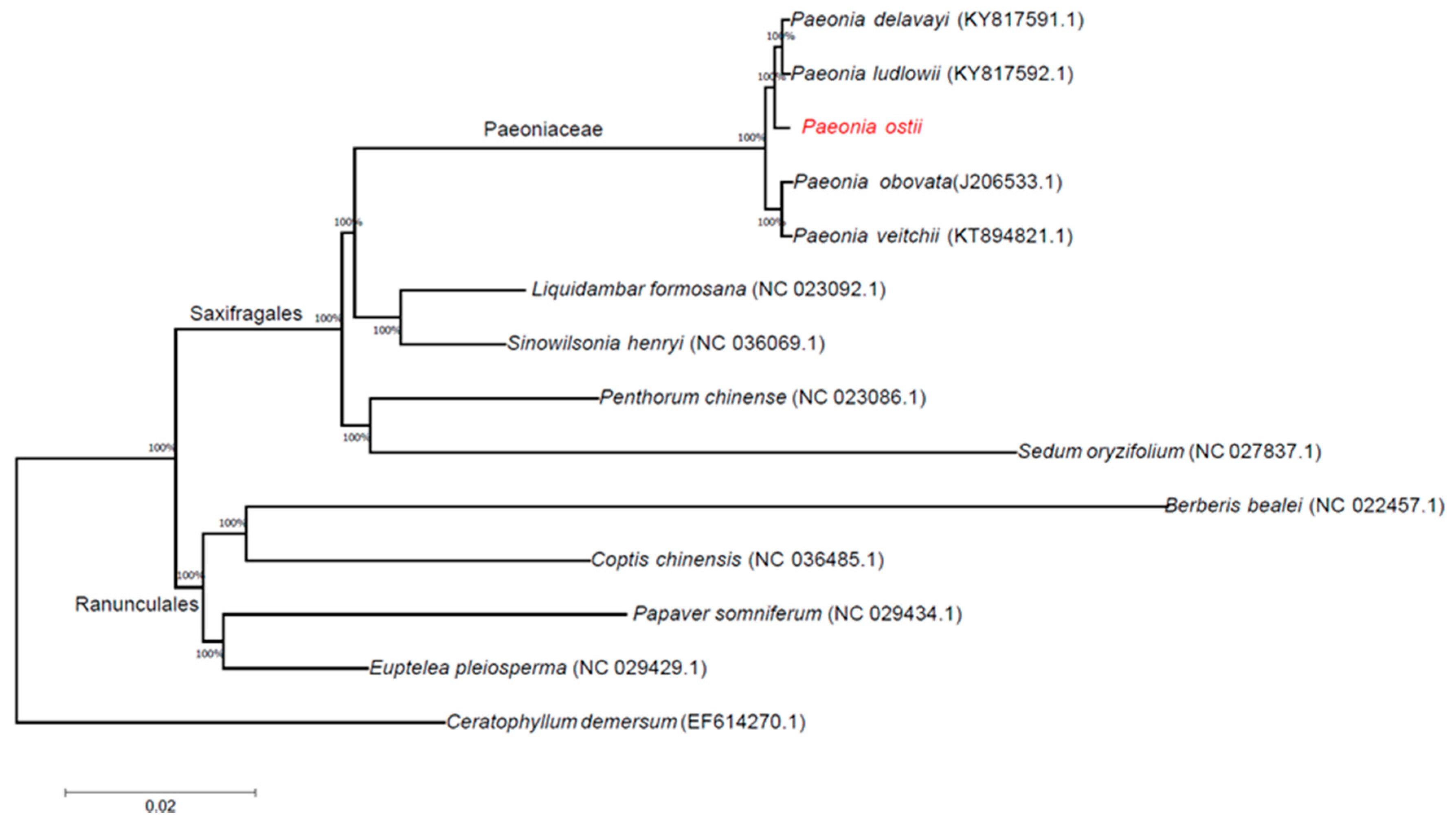
2.5. Phylogenetic Analysis
3. Materials and Methods
3.1. DNA Sequencing, Chloroplast Genome Assembly, and Validation
3.2. Gene Annotation and Sequence Analyses
3.3. Genome Comparison
3.4. Phylogenetic Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Wang, Y.; Dong, C.; Xue, Z.; Jin, Q.; Xu, Y. De novo transcriptome sequencing and discovery of genes related to copper tolerance in Paeonia ostii. Gene 2016, 576, 126–135. [Google Scholar] [CrossRef] [PubMed]
- Gai, S.; Zhang, Y.; Liu, C.G.S.; Zhang, Y.; Liu, C.; Zhang, Y. Transcript profiling of Paoenia ostii during artificial chilling induced dormancy release identifies activation of GA pathway and carbohydrate metabolism. PLoS ONE 2013, 8, e55297. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Song, Y.; Liu, Z.; Hu, Y. Culturable bacterial community analysis in the root domains of two varieties of tree peony (Paeonia ostii). FEMS Microbiol. Lett. 2011, 322, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.H.; Cheng, F.Y.; Zhou, S.L. The phylogeographic structure and conservation genetics of the endangered tree peony, Paeonia rockii (Paeoniaceae), inferred from chloroplast gene sequences. Conserv. Genet. 2011, 12, 1539–1549. [Google Scholar] [CrossRef]
- Deng, R.X.; Liu, Z.; Qin, L.L.; Wang, L.; Liu, X.Q.; Liu, P. Optimization of supercritical CO2 extraction and analysis of chemical composition of peony seed oil. Food Sci. 2010, 10, 142–145. [Google Scholar]
- Han, X.Y.; Zhang, Y.L.; Niu, L.X.; Luo, J.R.; Amp, N.A.; University, F. Fatty acid composition of ‘Fengdan‘ peony seed oils from different growing regions. Food Sci. 2014, 22, 181–184. [Google Scholar]
- Qi, J.C.; Zhou, H.M.; Ma, J.Q.; Pu, L. Analysis of the chemical constituents in peony seed oil by GC-MS. J. Cereals Oils 2005, 11, 22–23. [Google Scholar]
- Sevim, D.; Senol, F.S.; Gulpinar, A.R.; Orhan, I.E.; Kaya, E.; Kartal, M.; Sener, B. Discovery of potent in vitro neuroprotective effect of the seed extracts from seven Paeonia L. (peony) taxa and their fatty acid composition. Ind. Crops Prod. 2013, 49, 240–246. [Google Scholar] [CrossRef]
- Yi, J.P.; Zhu, W.X.; Ma, H.L.; Wang, Y.F. Optimization on ultrasonic-assisted extraction technology of oil from Paeonia suffruticosa Andr. seeds with response surface analysis. Trans. Chin. Soc. Agric. Mach. 2009, 40, 103–110. [Google Scholar]
- Zhang, X.; Zhang, Y.; Niu, L.; Sun, J.; Li, L.; Zhang, J.; Li, J. Chemometric classification of different tree peony species native to China based on the assessment of major fatty acids of seed oil and phenotypic characteristics of the seeds. Chem. Biodives. 2017, 14, 1612–1880. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.M.; Ma, J.Q.; Miao, C.Y.; Hu, J.L.; Yang, Z.Y.; Li, P.; Zheng, M.L. Physicochemical indexes and fatty acid composition of peony seed oil. China Oils Fats 2009, 34, 72–74. [Google Scholar]
- Barceló-Coblijn, G.; Murphy, E.J. Alpha-linolenic acid and its conversion to longer chain n−3 fatty acids: Benefits for human health and a role in maintaining tissue n−3 fatty acid levels. Prog. Lipid Res. 2009, 48, 355–374. [Google Scholar] [CrossRef] [PubMed]
- Carlson, S.J.; Fallon, E.M.; Kalish, B.T.; Gura, K.M.; Puder, M. The role of the ω-3 fatty acid DHA in the human life cycle. JPEN J. Parenter. Enteral Nutr. 2013, 37, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Stark, A.H.; Crawford, M.A.; Reifen, R. Update on alpha-linolenic acid. Nutr. Rev. 2008, 66, 326–332. [Google Scholar] [CrossRef] [PubMed]
- Lessard-Therrien, M.; Davies, T.J.; Bolmgren, K. A phylogenetic comparative study of flowering phenology along an elevational gradient in the Canadian subarctic. Int. J. Biometeorol. 2014, 58, 455–462. [Google Scholar] [CrossRef] [PubMed]
- Brunkard, J.O.; Runkel, A.M.; Zambryski, P.C. Chloroplasts extend stromules independently and in response to internal redox signals. Proc. Natl. Acad. Sci. USA 2015, 112, 10044–10049. [Google Scholar] [CrossRef] [PubMed]
- Daniell, H.; Chase, C. Molecular Biology and Biotechnology of Plant Organelles; Springer: Dordrecht, The Netherlands, 2004. [Google Scholar]
- Shen, X.; Wu, M.; Liao, B.; Liu, Z.; Bai, R.; Xiao, S.; Li, X.; Zhang, B.; Xu, J.; Chen, S. Complete chloroplast genome sequence and phylogenetic analysis of the medicinal plant Artemisia annua. Molecules 2017, 22, 1330. [Google Scholar] [CrossRef] [PubMed]
- Jansen, R.K.; Raubeson, L.A.; Boore, J.L.; Depamphilis, C.W.; Chumley, T.W.; Haberle, R.C.; Wyman, S.K.; Alverson, A.J.; Peery, R.; Herman, S.J. Methods for obtaining and analyzing whole chloroplast genome sequences. Methods Enzymol. 2005, 395, 348–384. [Google Scholar] [PubMed]
- Chen, X.; Liu, C. Progress in Chloroplast Genome Analysis. Prog. Biochem. Biophys. 2008, 35, 21–28. [Google Scholar]
- Tao, S.; Crawford, D.J.; Stuessy, T.F. Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae). Am. J. Bot. 1997, 84, 1120–1136. [Google Scholar]
- Yuan, J.H.; Cheng, F.Y.; Zhou, S.L. Hybrid origin of Paeonia x Yananensis revealed by microsatellite markers, chloroplast gene sequences, and morphological characteristics. Int. J. Plant Sci. 2010, 171, 409–420. [Google Scholar] [CrossRef]
- Zhang, J.M.; Liu, J.; Sun, H.L.; Yu, J.; Wang, J.X.; Zhou, S.L. Nuclear and chloroplast SSR markers in Paeonia delavayi (Paeoniaceae) and cross-species amplification in P. ludlowii. Am. J. Bot. 2011, 98, 346–348. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.M.; Liu, J.; Sun, H.L.; Yu, J.; Wang, J.X.; Zhou, S.L. Phylogenetic analysis of Paeonia sect. Moutan (Paeoniaceae) based on multiple DNA fragments and morphological data. J. Syst. Evol. 2008, 46, 563–572. [Google Scholar]
- Zhou, S.L.; Zou, X.H.; Zhou, Z.Q.; Liu, J.; Xu, C.; Yu, J.; Wang, Q.; Zhang, D.M.; Wang, X.Q.; Ge, S. Multiple species of wild tree peonies gave rise to the ‘king of flowers’, Paeonia suffruticosa Andrews. Proc. Biol. Sci. 2014, 281, 1687. [Google Scholar] [CrossRef] [PubMed]
- Sidonie, B.; Natalie, C.; Luo, S.; Sun, G.; Shahin, Z.; Andreas, G.; Eva, T.; Renner, S.S. Assembled plastid and mitochondrial genomes, as well as nuclear genes, place the parasite family Cynomoriaceae in the Saxifragales. Genome Biol. Evol. 2016, 8, 2214–2230. [Google Scholar]
- Asaf, S.; Khan, A.L.; Khan, M.A.; Waqas, M.; Kang, S.M.; Yun, B.W.; Lee, I.J. Chloroplast genomes of Arabidopsis halleri ssp. gemmifera and Arabidopsis lyrata ssp. petraea: Structures and comparative analysis. Sci. Rep. 2017, 7, 7556. [Google Scholar] [PubMed]
- Ng, P.K.; Lin, S.M.; Lim, P.E.; Liu, L.C.; Chen, C.M.; Pai, T.W. Complete chloroplast genome of Gracilaria firma (Gracilariaceae, Rhodophyta), with discussion on the use of chloroplast phylogenomics in the subclass Rhodymeniophycidae. BMC Genom. 2017, 18, 40. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Dong, W.; Li, W.; Lu, Y.; Xie, X.; Jin, X.; Shi, J.; He, K.; Suo, Z. Comparative analysis of six Lagerstroemia complete chloroplast genomes. Front. Plant Sci. 2017, 8, 15. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Xiao, H.; Deng, C.; Xiong, L.; Yang, J.; Peng, C. The complete chloroplast genome sequences of the medicinal plant Pogostemon cablin. Int. J. Mol. Sci. 2016, 17, 820. [Google Scholar] [CrossRef] [PubMed]
- Shetty, S.M.; Md Shah, M.U.; Makale, K.; Mohdyusuf, Y.; Khalid, N.; Othman, R.Y. Complete chloroplast genome sequence of Musa balbisiana corroborates structural heterogeneity of inverted repeats in wild progenitors of cultivated bananas and plantains. Plant Genome 2016, 9, 1940–3372. [Google Scholar] [CrossRef] [PubMed]
- Boudreau, E.; Takahashi, Y.; Lemieux, C.; Turmel, M.; Rochaix, J.D. The chloroplast ycf3 and ycf4 open reading frames of Chlamydomonas reinhardtii are required for the accumulation of the photosystem I complex. EMBO J. 1997, 16, 6095–6104. [Google Scholar] [CrossRef] [PubMed]
- Naver, H.; Boudreau, E.; Rochaix, J.D. Functional studies of Ycf3: Its role in assembly of photosystem I and interactions with some of its subunits. Plant Cell 2001, 13, 2731–2745. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Cui, L.; Depamphilis, C.W.; Moret, B.M.; Tang, J. Gene rearrangement analysis and ancestral order inference from chloroplast genomes with inverted repeat. BMC Genomics 2008, 9, S25. [Google Scholar]
- Haberle, R.C.; Fourcade, H.M.; Boore, J.L.; Jansen, R.K. Extensive rearrangements in the chloroplast genome of Trachelium caeruleum are associated with repeats and tRNA genes. J. Mol. Evol. 2008, 66, 350–361. [Google Scholar] [CrossRef] [PubMed]
- Downie, S.R.; Llanas, E.; Katz-Downie, D.S. Multiple independent losses of the rpoC1 intron in angiosperm chloroplast DNA’s. Syst. Bot. 1996, 21, 135–151. [Google Scholar] [CrossRef]
- Downie, S.R.; Olmstead, R.G.; Zurawski, G.; Soltis, D.E.; Soltis, P.S.; Watson, J.C.; Palmer, J.D. Six independent losses of the chloroplast DNA rpl2 intron in dicotyledons: Molecular and phylogenetic implications. Evolution 1991, 45, 1245–1259. [Google Scholar] [CrossRef] [PubMed]
- Graveley, B.R. Alternative splicing: Increasing diversity in the proteomic world. Trends Genet. 2001, 17, 100–107. [Google Scholar] [CrossRef]
- Guisinger, M.M.; Chumley, T.W.; Kuehl, J.V.; Boore, J.L.; Jansen, R.K. Implications of the plastid genome sequence of Typha (Typhaceae, Poales) for understanding genome evolution in Poaceae. J. Mol. Evol. 2010, 70, 149–166. [Google Scholar] [CrossRef] [PubMed]
- Jansen, R.K.; Cai, Z.; Raubeson, L.A.; Daniell, H.; Depamphilis, C.W.; Leebensmack, J.; Kai, F.M. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl. Acad. Sci. USA 2007, 104, 19369–19374. [Google Scholar] [CrossRef] [PubMed]
- Ueda, M.; Fujimoto, M.; Arimura, S.; Murata, J.; Tsutsumi, N.; Kadowaki, K. Loss of the rpl32 gene from the chloroplast genome and subsequent acquisition of a preexisting transit peptide within the nuclear gene in Populus. Gene 2007, 402, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Chu, Y.; Liao, B.; Xiao, S.; Yin, Q.; Bai, R.; Su, H.; Dong, L.; Li, X.; Qian, J.; et al. Panax ginseng genome examination for ginsenoside biosynthesis. Gigascience 2017, 6, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Niu, D.; Yang, Y. Why eukaryotic cells use introns to enhance gene expression: Splicing reduces transcription-associated mutagenesis by inhibiting topoisomerase I cutting activity. Biol. Direct 2011, 6, 24. [Google Scholar] [CrossRef] [PubMed]
- Le, H.; Nott, A.; Moore, M.J. How introns influence and enhance eukaryotic gene expression. Trends Biochem. Sci. 2003, 28, 215–220. [Google Scholar]
- Callis, J.; Fromm, M.; Walbot, V. Introns increase gene expression in cultured maize cells. Genes Dev. 1987, 1, 1183–1200. [Google Scholar] [CrossRef] [PubMed]
- Emami, S.; Arumainayagam, D.; Korf, I.; Rose, A.B. The effects of a stimulating intron on the expression of heterologous genes in Arabidopsis thaliana. Plant Biotechnol. J. 2013, 11, 555–563. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Xu, C.; Cheng, T.; Lin, K.; Zhou, S. Sequencing angiosperm plastid genomes made easy: A complete set of universal primers and a case study on the phylogeny of saxifragales. Genome Biol. Evol. 2013, 5, 989–997. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Xu, C.; Li, D.; Jin, X.; Li, R.; Lu, Q.; Suo, Z. Comparative analysis of the complete chloroplast genome sequences in psammophytic Haloxylon species (Amaranthaceae). Peer J. 2016, 4, e2699. [Google Scholar] [CrossRef] [PubMed]
- Suo, Z.; Li, W.Y.; Jin, X.B.; Zhang, H.J. A new nuclear DNA marker revealing both microsatellite variations and single nucleotide polymorphic loci: A case study on classification of cultivars in Lagerstroemia indica L. J. Microb. Biochem. Technol. 2016, 8, 266–271. [Google Scholar] [CrossRef]
- Yang, Y.; Zhou, T.; Duan, D.; Yang, J.; Feng, L.; Zhao, G. Comparative analysis of the complete chloroplast genomes of five Quercus species. Front. Plant Sci. 2016, 7, 959. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.H.; Xu, J.; Xiao, S.M.; Liao, B.S.; Gao, Y.; Zhai, C.C.; Qiu, X.H.; Xu, W.; Chen, S.L. Comparative optical genome analysis of two pangolin species: Manis pentadactyla and Manis javanica. Gigascience 2016, 5, 1–5. [Google Scholar]
- Chen, S.; Xu, J.; Liu, C.; Zhu, Y.; Nelson, D.R.; Zhou, S.; Li, C.; Wang, L.; Guo, X.; Sun, Y. Genome sequence of the model medicinal mushroom Ganoderma lucidum. Nat. Commun. 2012, 3, 913. [Google Scholar] [CrossRef] [PubMed]
- Ni, L.; Zhao, Z.; Xu, H.; Chen, S.; Dorje, G. The complete chloroplast genome of Gentiana straminea (Gentianaceae), an endemic species to the Sino-Himalayan subregion. Gene 2016, 577, 281–288. [Google Scholar] [CrossRef] [PubMed]
- Kode, V.; Mudd, E.A.; Iamtham, S.; Day, A. The tobacco plastid accD gene is essential and is required for leaf development. Plant J. 2005, 44, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Raubeson, L.A.; Peery, R.; Chumley, T.W.; Dziubek, C.; Fourcade, H.M.; Boore, J.L.; Jansen, R.K. Comparative chloroplast genomics: Analyses including new sequences from the angiosperms Nuphar advena and Ranunculus macranthus. BMC Genom. 2007, 8, 174. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Tang, P.; Li, Z.; Li, D.; Liu, Y.; Huang, H. The first complete chloroplast genome sequences in Actinidiaceae: Genome structure and comparative analysis. PLoS ONE 2015, 10, e0129347. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Xu, C.; Li, C.; Sun, J.; Zuo, Y.; Shi, S.; Cheng, T.; Guo, J.; Zhou, S. ycf1, the most promising plastid DNA barcode of land plants. Sci. Rep. 2015, 5, 8348. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, S.; Bédard, J.; Hirano, M.; Hirabayashi, Y.; Oishi, M.; Imai, M.; Takase, M.; Ide, T.; Nakai, M. Uncovering the protein translocon at the chloroplast inner envelope membrane. Science 2013, 339, 571–574. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.J.; Davis, J.I.; Soreng, R.J.; Garvin, D.; Anderson, M.J. Chloroplast DNA inversions and the origin of the grass family (Poaceae). Proc. Natl. Acad. Sci. USA 1992, 89, 7722–7726. [Google Scholar] [CrossRef] [PubMed]
- Jansen, R.K.; Palmer, J.D. A chloroplast DNA inversion marks an ancient evolutionary split in the sunflower family (Asteraceae). Proc. Natl. Acad. Sci. USA 1987, 84, 5818–5822. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Hahn, F.M.; Mcmahan, C.M.; Cornish, K.; Whalen, M.C. Comparative analysis of the complete sequence of the plastid genome of Parthenium argentatum and identification of DNA barcodes to differentiate Parthenium species and lines. BMC Plant Biol. 2009, 9, 131. [Google Scholar] [CrossRef] [PubMed]
- Palmer, J.D.; Nugent, J.M.; Herbon, L.A. Unusual structure of geranium chloroplast DNA: A triple-sized inverted repeat, extensive gene duplications, multiple inversions, and two repeat families. Proc. Natl. Acad. Sci. USA 1987, 84, 769–773. [Google Scholar] [CrossRef] [PubMed]
- Raubeson, L.A.; Jansen, R.K. Chloroplast DNA evidence on the ancient evolutionary split in vascular land plants. Science 1992, 255, 1697–1699. [Google Scholar] [CrossRef] [PubMed]
- Shi, Q.H.; Yao, Z.P.; Zhang, H.; Xu, L.; Dai, P.H. Comparison of four methods of DNA extraction from chickpea. J. Xinjiang Agric. Univ. 2009, 1, 64–67. [Google Scholar]
- Jiang, H.; Rong, L.; Ding, S.W.; Zhu, S. Skewer: A fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinform. 2014, 15, 182. [Google Scholar] [CrossRef] [PubMed]
- Deng, P.C.; Wang, L.; Cui, L.C.; Feng, K.W.; Liu, F.Y.; Du, X.D.; Tong, W.; Nie, X.J.; Ji, W.Q.; Weining, S. Global identification of microRNAs and their targets in barley under salinity stress. PLoS ONE 2015, 10, e0137990. [Google Scholar] [CrossRef] [PubMed]
- Gogniashvili, M.; Naskidashvili, P.; Bedoshvili, D.; Kotorashvili, A.; Kotaria, N.; Beridze, T. Complete chloroplast DNA sequences of Zanduri wheat ( Triticum spp.). Genet. Resour. Crop Evol. 2015, 62, 1269–1277. [Google Scholar] [CrossRef]
- Acemel, R.D.; Tena, J.J.; Irastorzaazcarate, I.; Marlétaz, F.; Gómezmarín, C.; Callemustienes, E.D.L.; Bertrand, S.; Diaz, S.G.; Aldea, D.; Aury, J.M. A single three-dimensional chromatin compartment in amphioxus indicates a stepwise evolution of vertebrate Hox bimodal regulation. Nat. Genet. 2016, 48, 336–341. [Google Scholar] [CrossRef] [PubMed]
- Boetzer, M.; Henkel, C.V.; Jansen, H.J.; Butler, D.; Pirovano, W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 2011, 27, 578–579. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Shi, L.; Zhu, Y.; Chen, H.; Zhang, J.; Lin, X.; Guan, X. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genom. 2012, 13, 715. [Google Scholar]
- Wyman, S.K.; Jansen, R.K.; Boore, J.L. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 2004, 20, 3252–3255. [Google Scholar] [CrossRef] [PubMed]
- Lohse, M.; Drechsel, O.; Bock, R. OrganellarGenomeDRAW (OGDRAW): A tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 2007, 52, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Frazer, K.A.; Pachter, L.; Poliakov, A.; Rubin, E.M.; Dubchak, I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004, 32, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Mayor, C.; Brudno, M.; Schwartz, J.R.; Poliakov, A.; Rubin, E.M.; Frazer, K.A.; Pachter, L.S.; Dubchak, I. VISTA: Visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 2000, 16, 1046–1047. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, S.; Choudhuri, J.V.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef] [PubMed]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Kraemer, L.; Beszteri, B.; Gäbler-Schwarz, S.; Held, C.; Leese, F.; Mayer, C.; Pöhlmann, K.; Frickenhau, S. STAMP: Extensions to the STADEN sequence analysis package for high throughput interactive microsatellite marker design. BMC Bioinform. 2009, 10, 41. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.; Blackshields, G.; Brown, N.; Chenna, R.; Mcgettigan, P.; Mcwilliam, H.; Valentin, F.; Wallace, I.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X v. 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Sequence data of Paeonia ostii is available from the authors. |
| Species | P. delavayi | P. ludlowii | P. obovata | P. veitchii | P. ostii |
|---|---|---|---|---|---|
| LSC (large single-copy) | |||||
| Length (bp) | 86,057 | 84,426 | 84,387 | 84,398 | 85,373 |
| G+C (%) | 36.7 | 36.7 | 36.7 | 36.7 | 36.7 |
| Length (%) | 56.4 | 55.3 | 55.3 | 55.3 | 56.1 |
| SSC (small single-copy) | |||||
| Length (bp) | 17,050 | 16,983 | 17,028 | 16,978 | 17,054 |
| G+C (%) | 32.7 | 32.7 | 32.7 | 32.7 | 32.7 |
| Length (%) | 11.2 | 11.1 | 11.2 | 11.1 | 11.2 |
| IR (inverse repeats) | |||||
| Length (bp) | 25,649 | 25,639 | 26,642 | 25,653 | 24,863 |
| G+C (%) | 43.1 | 43.1 | 43.1 | 43.1 | 43.1 |
| Length (%) | 16.8 | 16.8 | 17.4 | 16.8 | 16.3 |
| Total | |||||
| Length (bp) | 152,698 | 152,687 | 152,698 | 152,682 | 152,153 |
| G+C (%) | 38.4 | 38.4 | 38.4 | 38.4 | 38.4 |
| Category | Group of Genes | Name of Genes |
|---|---|---|
| Self-replication | Large subunit of ribosomal proteins | rpl2 *,a, 14, 16 *, 20, 22, 23 a, 33, 36 |
| Small subunit of ribosomal proteins | rps2, 3, 4, 7 a, 8, 11, 12 *,a, 14, 16 *, 18, 19 | |
| DNA-dependent RNA polymerase | rpoA, B, C1 *, C2 | |
| rRNA genes | rrn16S a, rrn23S a, rrn4.5S a, rrn5S a | |
| tRNA genes | trnA-UGC *,a, trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnfM-CAU, trnG-UCC *, trnG-GCC, trnH-GUG, trnI-CAU, trnI-GAU *,a, trnK-UUU *, trnL-CAA, trnL-UAA *, trnL-UAG, trnM-CAU, trnN-GUU, trnP-UGG, trnQ-UUG, trnR-ACG, trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC, trnV-UAC *, trnW-CCA, trnY-GUA | |
| Photosynthesis | Photosystem I | psaA, B, C, I, J |
| Photosystem II | psbA, B, C, D, E, F, H, I, J, K, L, M, N, T, Z, | |
| NADH oxidoreductase | ndhA *, B *,a, C, D, E, F, G, H, I, J, K | |
| Cytochrome b6/f complex | petA, B *, D *, G, L, N | |
| ATP synthase | atpA, B, E, F *, H, I | |
| Rubisco | rbcL | |
| Other genes | Maturase | matK |
| Protease | clpP * | |
| Envelope membrane protein | cemA | |
| Subunit acetyl-CoA-carboxylase | accD | |
| c-Type cytochrome synthesis gene | ccsA | |
| Conserved open reading frames | ycf1, 2 a, 3 *, 4, 15 |
| Amino Acid | Codon | No. | RSCU * | tRNA | Amino Acid | Codon | No. | RSCU * | tRNA |
|---|---|---|---|---|---|---|---|---|---|
| Phe | UUU | 1226 | 1.16 | Tyr | UAU | 937 | 1.36 | ||
| Phe | UUC | 885 | 0.84 | trnF-GAA | Tyr | UAC | 440 | 0.64 | trnY-GUA |
| Leu | UUA | 769 | 1.24 | trnL-UAA | Stop | UAA | 683 | 1.1 | |
| Leu | UUG | 831 | 1.34 | trnL-CAA | Stop | UAG | 604 | 0.97 | |
| Leu | CUU | 690 | 1.11 | His | CAU | 486 | 1.34 | ||
| Leu | CUC | 483 | 0.78 | His | CAC | 240 | 0.66 | trnH-GUG | |
| Leu | CUA | 563 | 0.91 | trnL-UAG | Gln | CAA | 703 | 1.27 | trnQ-UUG |
| Leu | CUG | 392 | 0.63 | Gln | CAG | 401 | 0.73 | ||
| Ile | AUU | 1055 | 1.19 | Asn | AAU | 1033 | 1.33 | ||
| Ile | AUC | 746 | 0.84 | trnI-GAU | Asn | AAC | 520 | 0.67 | trnN-GUU |
| Ile | AUA | 855 | 0.97 | trnI-UAU | Lys | AAA | 1334 | 1.24 | trnK-UUU |
| Met | AUG | 743 | 1 | trn(f)M-CAU | Lys | AAG | 817 | 0.76 | |
| Val | GUU | 564 | 1.27 | Asp | GAU | 705 | 1.46 | ||
| Val | GUC | 293 | 0.66 | trnV-GAC | Asp | GAC | 264 | 0.54 | trnD-GUC |
| Val | GUA | 548 | 1.24 | trnV-UAC | Glu | GAA | 923 | 1.27 | trnE-UUC |
| Val | GUG | 366 | 0.83 | Glu | GAG | 526 | 0.73 | ||
| Ser | UCU | 659 | 1.45 | Cys | UGU | 393 | 1.19 | ||
| Ser | UCC | 458 | 1.01 | trnS-GGA | Cys | UGC | 270 | 0.81 | trnC-GCA |
| Ser | UCA | 599 | 1.32 | trnS-UGA | Stop | UGA | 580 | 0.93 | |
| Ser | UCG | 388 | 0.85 | Trp | UGG | 626 | 1 | trnW-CCA | |
| Pro | CCU | 368 | 1.02 | Arg | CGU | 217 | 0.58 | trnR-ACG | |
| Pro | CCC | 309 | 0.86 | trnP-GGG | Arg | CGC | 182 | 0.49 | |
| Pro | CCA | 427 | 1.18 | trnP-UGG | Arg | CGA | 400 | 1.07 | |
| Pro | CCG | 341 | 0.94 | Arg | CGG | 372 | 1 | ||
| Thr | ACU | 382 | 1.15 | Arg | AGA | 330 | 0.73 | trnR-UCU | |
| Thr | ACC | 290 | 0.87 | trnT-GGU | Arg | AGG | 291 | 0.64 | |
| Thr | ACA | 407 | 1.23 | trnT-UGU | Ser | AGU | 632 | 1.69 | |
| Thr | ACG | 248 | 0.75 | Ser | AGC | 439 | 1.17 | trnS-GCU | |
| Ala | GCU | 294 | 1.19 | Gly | GGU | 394 | 0.9 | ||
| Ala | GCC | 227 | 0.92 | Gly | GGC | 261 | 0.6 | trnG-GCC | |
| Ala | GCA | 253 | 1.02 | trnA-UGC | Gly | GGA | 579 | 1.33 | trnG-UCC |
| Ala | GCG | 217 | 0.88 | Gly | GGG | 509 | 1.17 |
| Gene | Location | Exon I (bp) | Intron I (bp) | Exon II (bp) | Intron II (bp) | Exon III (bp) |
|---|---|---|---|---|---|---|
| trnK-UUU | LSC | 35 | 2452 | 38 | ||
| trnG-UCC | LSC | 23 | 709 | 49 | ||
| trnL-UAA | LSC | 37 | 522 | 51 | ||
| trnV-UAC | LSC | 38 | 576 | 39 | ||
| trnI-GAU | IR | 42 | 935 | 36 | ||
| trnA-UGC | IR | 37 | 42 | 29 | ||
| rps12 * | LSC | 26 | 543 | 227 | 114 | |
| rps16 | LSC | 234 | 820 | 40 | ||
| rpl16 | LSC | 402 | 1008 | 10 | ||
| rpl2 | IR | 435 | 670 | 394 | ||
| rpoC1 | LSC | 1617 | 709 | 436 | ||
| ndhA | SSC | 540 | 1013 | 544 | ||
| ndhB | IR | 756 | 684 | 778 | ||
| ycf3 | SSC | 153 | 765 | 229 | 721 | 126 |
| petB | LSC | 6 | 754 | 658 | ||
| atpF | LSC | 408 | 701 | 160 | ||
| clpP | LSC | 228 | 659 | 292 | 673 | 67 |
| petD | LSC | 9 | 645 | 526 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, S.; Guo, L.; Zhao, W.; Xu, J.; Li, Y.; Zhang, X.; Shen, X.; Wu, M.; Hou, X. Complete Chloroplast Genome Sequence and Phylogenetic Analysis of Paeonia ostii. Molecules 2018, 23, 246. https://doi.org/10.3390/molecules23020246
Guo S, Guo L, Zhao W, Xu J, Li Y, Zhang X, Shen X, Wu M, Hou X. Complete Chloroplast Genome Sequence and Phylogenetic Analysis of Paeonia ostii. Molecules. 2018; 23(2):246. https://doi.org/10.3390/molecules23020246
Chicago/Turabian StyleGuo, Shuai, Lili Guo, Wei Zhao, Jiang Xu, Yuying Li, Xiaoyan Zhang, Xiaofeng Shen, Mingli Wu, and Xiaogai Hou. 2018. "Complete Chloroplast Genome Sequence and Phylogenetic Analysis of Paeonia ostii" Molecules 23, no. 2: 246. https://doi.org/10.3390/molecules23020246