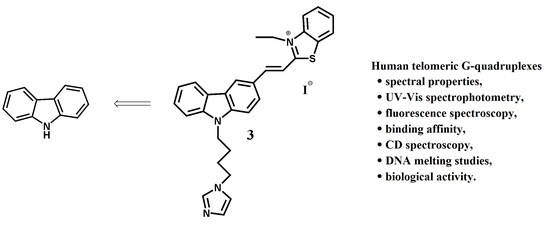
Binding Study of the Fluorescent Carbazole Derivative with Human Telomeric G-Quadruplexes
Abstract
:1. Introduction
2. Results and Discussion
2.1. Chemistry
2.2. Spectral Properties of Ligand 3
2.3. Spectrophotometric Titration
2.4. Fluorescence Spectroscopy
2.5. Binding Parameters of Ligand/22HT Complex
2.6. Circular Dichroism (CD) Spectroscopy
2.7. DNA Melting Studies
2.8. Biological Activity
3. Conclusions
4. Experimental
4.1. Materials
4.1.1. Ligands
General Procedure for Synthesis of Carbazole Derivatives
4.1.2. Oligonucleotide
4.2. Methods
4.2.1. Absorption Spectroscopy
4.2.2. Continuous Variation Analysis
4.2.3. Fluorescence Spectroscopy
4.2.4. Circular Dichroism
4.2.5. Ligand 3—22HT G4 Binding Study
4.2.6. Biological Activity
Cytotoxic effect–MTT assay
Telomerase Repeat Amplification Protocol (TRAP) Assay
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Burge, S.; Parkinson, G.N.; Hazel, P.; Todd, A.K.; Neidle, S. Quadruplex DNA: Sequence, topology and structure. Nucleic Acids Res. 2006, 34, 5402–5415. [Google Scholar] [CrossRef] [PubMed]
- De Cian, A.; Lacroix, L.; Douarre, C.; Temime-Smaali, N.; Trentesaux, C.; Riou, J.-F.; Mergny, J.-L. Targeting telomeres and telomerase. Biochimie 2008, 90, 131–155. [Google Scholar] [CrossRef] [PubMed]
- Ou, T.-M.; Lu, Y.-J.; Tan, J.-H.; Huang, Z.-S.; Wong, K.-Y.; Gu, L.-Q. G-quadruplexes: Targets in anticancer drug design. ChemMedChem 2008, 3, 690–713. [Google Scholar] [CrossRef]
- Folini, M.; Gandellini, P.; Zaffaroni, N. Targeting the telosome: Therapeutic implications. Biochim. Biophys. Acta 2009, 1792, 309–316. [Google Scholar] [CrossRef] [Green Version]
- Biffi, G.; Tannahill, D.; McCafferty, J.; Balasubramanian, S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013, 5, 182–186. [Google Scholar] [CrossRef] [Green Version]
- Biffi, G.; Di Antonio, M.; Tannahill, D.; Balasubramanian, S. Visualization and selective chemical targeting of RNA Gquadruplex structures in the cytoplasm of human cells. Nat. Chem. 2014, 6, 75–80. [Google Scholar] [CrossRef]
- Murat, P.; Balasubramanian, S. Existence and consequences of G-quadruplex structures in DNA. Curr. Opin. Genet. Dev. 2014, 25, 22–29. [Google Scholar] [CrossRef]
- Rhodes, D.; Lipps, H.J. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015, 43, 8627–8637. [Google Scholar] [CrossRef] [Green Version]
- Simonsson, T.; Pecinka, P.; Kubista, M. DNA tetraplex formation in the control region of c-myc. Nucleic Acids Res. 1998, 26, 1167–1172. [Google Scholar] [CrossRef] [Green Version]
- Phan, A.T.; Modi, Y.S.; Patel, D.J. Propeller-Type Parallel-Stranded G-Quadruplexes in the Human c-myc Promoter. J. Am. Chem. Soc. 2004, 126, 8710–8716. [Google Scholar] [CrossRef]
- Rankin, S.; Reszka, A.P.; Huppert, J.; Zloh, M.; Parkinson, G.N.; Todd, A.K.; Ladame, S.; Balasubramanian, S.; Neidle, S. Putative DNA Quadruplex Formation within the Human c-kit Oncogene. J. Am. Chem. Soc. 2005, 127, 10584–10589. [Google Scholar] [CrossRef] [PubMed]
- Todd, A.K.; Haider, S.M.; Parkinson, G.N.; Neidle, S. Sequence occurrence and structural uniqueness of a G-quadruplex in the human c-kit promoter. Nucleic Acids Res. 2007, 35, 5799–5808. [Google Scholar] [CrossRef] [PubMed]
- Dexheimer, T.S.; Sun, D.; Hurley, L.H. Deconvoluting the Structural and Drug-Recognition Complexity of the G-Quadruplex-Forming Region Upstream of the bcl-2 P1 Promoter. J. Am. Chem. Soc. 2006, 128, 5404–5415. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, P.; Lin, C.; Mathad, R.I.; Carver, M.; Yang, D. The Major G-Quadruplex Formed in the Human BCL-2 Proximal Promoter Adopts a Parallel Structure with a 13-nt Loop in K+ Solution. J. Am. Chem. Soc. 2014, 136, 1750–1753. [Google Scholar] [CrossRef] [PubMed]
- Guo, K.; Pourpak, A.; Beetz-Rogers, K.; Gokhale, V.; Sun, D.; Hurley, L.H. Formation of Pseudosymmetrical G-Quadruplex and i-Motif Structures in the Proximal Promoter Region of the RET Oncogene. J. Am. Chem. Soc. 2007, 129, 10220–10228. [Google Scholar] [CrossRef] [PubMed]
- Tong, X.; Lan, W.; Zhang, X.; Wu, H.; Liu, M.; Cao, C. Solution structure of all parallel G-quadruplex formed by the oncogene RET promoter sequence. Nucleic Acids Res. 2011, 39, 6753–6763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agrawal, P.; Hatzakis, E.; Guo, K.; Carver, M.; Yang, D. Solution structure of the major G-quadruplex formed in the human VEGF promoter in K+: Insights into loop interactions of the parallel G-quadruplexes. Nucleic Acids Res. 2013, 41, 10584–10592. [Google Scholar] [CrossRef]
- Parkinson, G.N.; Lee, M.P.H.; Neidle, S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature 2002, 417, 876–880. [Google Scholar] [CrossRef]
- Harley, C.B.; Futcher, A.B.; Greider, C.W. Telomeres shorten during ageing of human fibroblasts. Nature 1990, 345, 458–460. [Google Scholar] [CrossRef]
- Blackburn, E.H. Structure and function of telomeres. Nature 1991, 350, 569–573. [Google Scholar] [CrossRef]
- Lowden, M.R.; Flibotte, S.; Moerman, D.G.; Ahmed, S. DNA synthesis generates terminal duplications that seal end-to-end chromosome fusions. Science 2011, 332, 468–471. [Google Scholar] [CrossRef]
- Meyne, J.; Ratliff, R.L.; Moyzis, R.K. Conservation of the human telomere sequence (TTAGGG)n among vertebrates. Proc. Natl. Acad. Sci. USA 1989, 86, 7049–7053. [Google Scholar] [CrossRef] [PubMed]
- Zahler, A.M.; Williamson, J.R.; Cech, T.R.; Prescott, D.M. Inhibition of telomerase by G-quartet DNA structures. Nature 1991, 350, 718–720. [Google Scholar] [CrossRef]
- Sun, D.; Thompson, B.; Cathers, B.E.; Salazar, M.; Kerwin, S.M.; Trent, J.O.; Jenkins, T.C.; Neidle, S.; Hurley, L.H. Inhibition of human telomerase by a G-quadruplex-interactive compound. J. Med. Chem. 1997, 40, 2113–2116. [Google Scholar] [CrossRef] [PubMed]
- Haudecoeur, R.; Stefan, L.; Denat, F.; Monchaud, D. A Model of Smart G Quadruplex Ligand. J. Am. Chem. Soc. 2013, 135, 550–553. [Google Scholar] [CrossRef] [PubMed]
- Ohnmacht, S.A.; Neidle, S. Small-molecule quadruplex-targeted drug discovery. Bioorg. Med. Chem. Lett. 2014, 24, 2602–2612. [Google Scholar] [CrossRef]
- Islam, M.M.; Fujii, S.; Sato, S.; Okauchi, T.; Takenaka, S. A Selective G-Quadruplex DNA-Stabilizing Ligand Based on a Cyclic Naphthalene Diimide Derivative. Molecules 2015, 20, 10963–10979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajczak, E.; Gluszynska, A.; Juskowiak, B. Interaction of metallacrown complexes with G-quadruplex DNA. J. Inorg. Biochem. 2016, 155, 105–114. [Google Scholar] [CrossRef]
- Rajczak, E.; Pecoraro, V.L.; Juskowiak, B. Sm(III)[12-MCGa(III)shi-4] as a luminescent probe for G-quadruplex structures. Metallomics 2017, 9, 1735–1744. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.-H.; Chen, S.-B.; Wang, B.; Ou, T.-M.; Gu, L.-Q.; Tan, J.-H.; Huang, Z.-S. Specific targeting of telomeric multimeric G-quadruplexes by a new triaryl-substituted imidazole. Nucleic Acids Res. 2017, 45, 1606–1618. [Google Scholar] [CrossRef]
- Islam, M.M.; Sato, S.; Shinozaki, S.; Takenaka, S. Cyclic ferrocenylnaphthalene diimide derivative as a new class of G-quadruplex DNA binding ligand. Bioorg. Med. Chem. Lett. 2017, 27, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Monsen, R.C.; Trent, J.O. G-quadruplex virtual drug screening: A review. Biochimie 2018, 152, 134–148. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-Y.; Wang, Q.; Liu, J.-Q.; Hao, Y.-H.; Tan, Z. Contribution of Telomere G-Quadruplex Stabilization to the Inhibition of Telomerase-Mediated Telomere Extension by Chemical Ligands. J. Am. Chem. Soc. 2011, 133, 15036–15044. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.-C.; Chu, J.-F.; Tsai, Y.-L.; Wang, Z.-F. Structure conversion and structure separation of G-quadruplexes investigated by carbazole derivatives. Curr. Pharm. Des. 2012, 18, 2002–2013. [Google Scholar] [CrossRef] [PubMed]
- Tseng, T.-Y.; Wang, Z.-F.; Chien, C.-H.; Chang, T.-C. In-cell optical imaging of exogenous G-quadruplex DNA by fluorogenic ligands. Nucleic Acids Res. 2013, 41, 10605–10618. [Google Scholar] [CrossRef]
- Tseng, T.-Y.; Chang, C.-C.; Lin, J.-J.; Chang, T.-C. A Fluorescent Anti-Cancer Agent, 3,6-bis(1-methyl-4-vinylpyridinium) Carbazole Diiodide, Stains G-Quadruplexes in Cells and Inhibits Tumor Growth. Curr. Top. Med. Chem. 2015, 15, 1964–1970. [Google Scholar] [CrossRef]
- Maji, B.; Kumar, K.; Kaulage, M.; Muniyappa, K.; Bhattacharya, S. Design and Synthesis of New Benzimidazole−Carbazole Conjugates for the Stabilization of Human Telomeric DNA, Telomerase Inhibition, and Their Selective Action on Cancer Cells. J. Med. Chem. 2014, 57, 6973–6988. [Google Scholar] [CrossRef] [PubMed]
- Maji, B.; Kumar, K.; Muniyappa, K.; Bhattacharya, S. New dimeric carbazole–benzimidazole mixed ligands for the stabilization of human telomeric G-quadruplex DNA and as telomerase inhibitors. A remarkable influence of the spacer. Org. Biomol. Chem. 2015, 13, 8335–8348. [Google Scholar] [CrossRef]
- Kaulage, M.H.; Maji, B.; Pasadi, S.; Ali, A.; Bhattacharya, S.; Muniyappa, K. Targeting G-quadruplex DNA structures in the telomere and oncogene promoter regions by benzimidazole‒carbazole ligands. Eur. J. Med. Chem. 2018, 148, 178–194. [Google Scholar] [CrossRef]
- Petraccone, L.; Fotticchia, I.; Cummaro, A.; Pagano, B.; Ginnari-Satriani, L.; Haider, S.; Randazzo, A.; Novellino, E.; Neidle, S.; Giancola, C. The triazatruxene derivative azatrux binds to the parallel form of the human telomeric G-quadruplex under molecular crowding conditions: Biophysical and molecular modeling studies. Biochimie 2011, 93, 1318–1327. [Google Scholar] [CrossRef]
- Sengupta, P.; Chattopadhyay, S.; Chatterjee, S. G-Quadruplex surveillance in BCL-2 gene: A promising therapeutic intervention in cancer treatment. Drug Discov. Today. 2017, 22, 1165–1186. [Google Scholar] [CrossRef] [PubMed]
- Debnath, M.; Paul, R.; Panda, D.; Dash, J. Enzyme-Regulated DNA-Based Logic Device. ACS Synth. Biol. 2018, 7, 1456–1464. [Google Scholar] [CrossRef] [PubMed]
- Głuszyńska, A.; Juskowiak, B.; Kuta-Siejkowska, M.; Hoffmann, M.; Haider, S. Carbazole ligands as c-myc G-quadruplex binders. Int. J. Biol. Macromol. 2018, 114, 479–490. [Google Scholar] [CrossRef] [PubMed]
- Głuszyńska, A.; Juskowiak, B.; Kuta-Siejkowska, M.; Hoffmann, M.; Haider, S. Carbazole derivatives’ binding to c-KIT G-quadruplex DNA. Molecules 2018, 23, 1134. [Google Scholar] [CrossRef] [PubMed]
- Głuszyńska, A.; Rajczak, E.; Juskowiak, B. Synthesis and spectroscopic characterisation of (E)-2-(2-(9-(4-(1H-1,2,4-triazol-1-yl)butyl)-9H-carbazol-3-yl)vinyl)-3-ethylbenzo- [d]thiazol-3-ium, a new ligand and potential DNA intercalator. Chem. Papers 2013, 67, 1231–1239. [Google Scholar] [CrossRef]
- Thomas, K.R.J.; Lin, J.T.; Tao, Y.-T.; Ko, C.-W. Novel green light-emitting carbazole derivatives: Potential electroluminescent materials. Adv Mater. 2000, 12, 1949–1951. [Google Scholar] [CrossRef]
- Agarwal, N.; Nayak, P.K.; Ali, F.; Patankar, M.P.; Narasimhan, K.L.; Periasamy, N. Tuning of HOMO levels of carbazole derivatives: New molecules for blue OLED. Synth. Met. 2011, 161, 466–473. [Google Scholar] [CrossRef]
- Manickam, M.; Iqbal, P.; Belloni, M.; Kumar, S.; Preece, J.A. A brief review of carbazole-based photorefractive liquid crystalline materials. Isr. J. Chem. 2012, 52, 917–934. [Google Scholar] [CrossRef]
- Venkateswararao, A.; Thomas, K.R.J. Carbazole-based organic dyes for dye-sensitized solar cells: Role of carbazole as donor, auxiliary donor and π-linker. Sol. Cell Nanotechnol. 2013, 41–96. [Google Scholar] [CrossRef]
- Reig, M.; Gozalvez, C.; Bujaldon, R.; Bagdziunas, G.; Ivaniuk, K.; Kostiv, N.; Volyniuk, D.; Grazulevicius, J.V.; Velasco, D. Easy accessible blue luminescent carbazole-based materials for organic light-emitting diodes. Dyes Pigm. 2017, 137, 24–35. [Google Scholar] [CrossRef]
- Zhu, L.; Shan, Y.; Wang, R.; Liu, D.; Zhong, C.; Song, Q.; Wu, F. High-efficiency perovskite solar cells based on new TPE compounds as hole transport materials: The role of 2,7- and 3,6-substituted carbazole derivatives. Chem. Eur. J. 2017, 23, 4373–4379. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, A.W.; Reddy, K.R.; Knölker, H.-J. Occurrence, Biogenesis, and Synthesis of Biologically Active Carbazole Alkaloids. Chem. Rev. 2012, 112, 3193–3328. [Google Scholar] [CrossRef] [PubMed]
- Głuszyńska, A. Biological potential of carbazole derivatives. Eur. J. Med. Chem. 2015, 94, 405–426. [Google Scholar] [CrossRef] [PubMed]
- Tsutsumi, L.S.; Gündisch, D.; Sun, D. Carbazole scaffold in medicinal chemistry and natural products: A review from 2010-2015. Curr. Top. Med. Chem. 2016, 16, 1290–1313. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Peng, X.-M.; Damu, G.L.V.; Geng, R.-X.; Zhou, C.-H. Comprehensive review in current developments of imidazole-based medicinal chemistry. Med. Res. Rev. 2014, 34, 340–437. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Kumar, V.; Kharb, R.; Kumar, S.; Sharma, P.C.; Pathak, D.P. Imidazole derivatives as potential therapeutic agents. Curr. Pharm. Des. 2016, 22, 3265–3301. [Google Scholar] [CrossRef] [PubMed]
- Ali, I.; Lone, M.N.; Aboul-Enein, H.Y. Imidazoles as potential anticancer agents. MedChemComm 2017, 8, 1742–1773. [Google Scholar] [CrossRef] [PubMed]
- Shalmali, N.; Ali, M.R.; Bawa, S. Imidazole: An essential edifice for the identification of new lead compounds and drug development. Mini Rev. Med. Chem. 2018, 18, 142–163. [Google Scholar] [CrossRef]
- Fan, Y.-L.; Jin, X.-H.; Huang, Z.-P.; Yu, H.-F.; Zeng, Z.-G.; Gao, T.; Feng, L.-S. Recent advances of imidazole-containing derivatives as anti-tubercular agents. Eur. J. Med. Chem. 2018, 150, 347–365. [Google Scholar] [CrossRef]
- Wang, Y.; Patel, D.J. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure 1993, 4, 263–282. [Google Scholar] [CrossRef]
- Ambrus, A.; Chen, D.; Dai, J.; Bialis, T.; Jones, R.A.; Yang, D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006, 34, 2723–2735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, J.; Carver, M.; Punchihewa, C.; Jones, R.A.; Yang, D. Structure of the Hybrid-2 type intramolecular human telomeric G-quadruplex in K+ solution: Insights into structure polymorphism of the human telomeric sequence. Nucleic Acids Res. 2007, 35, 4927–4940. [Google Scholar] [CrossRef] [PubMed]
- Luu, K.N.; Phan, A.T.; Kuryavyi, V.; Lacroix, L.; Patel, D.J. Structure of the human telomere in K+ solution: An intramolecular (3 + 1) G-quadruplex scaffold. J. Am. Chem. Soc. 2006, 128, 9963–9970. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Noguchi, Y.; Sugiyama, H. The new models of the human telomere d[AGGG(TTAGGG)3] in K+ solution. Bioorg. Med. Chem. 2006, 14, 5584–5591. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.-F.; Gan, L.-L.; Zhou, C.-H. Synthesis, antibacterial and antifungal activities of some carbazole derivatives. Bioorg. Med. Chem. Lett. 2010, 20, 1881–1884. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, L.; Wada, T.; Sasabe, H. Synthesis and characterization of novel hyperbranched polymer with dipole carbazole moieties for multifunctional materials. J. Polym. Sci. A Polym. Chem. 1996, 34, 1359–1363. [Google Scholar] [CrossRef] [Green Version]
- Xu, T.H.; Lu, R.; Qiu, X.P.; Liu, X.L.; Xue, P.C.; Tan, C.H.; Bao, C.Y.; Zhao, Y.Y. Synthesis and characterization of carbazole-based dendrimers with porphyrin cores. Eur. J. Org. Chem. 2006, 2006, 4014–4020. [Google Scholar] [CrossRef]
- Ryu, H.; Subramanian, L.R.; Hanack, M. Photoand electroluminescent properties of cyano-substituted styryl derivatives and synthesis of CN–PPV model compounds containing an alkoxy spacer for OLEDs. Tetrahedron 2006, 62, 6236–6247. [Google Scholar] [CrossRef]
- Song, Y.; Di, C.A.; Wei, Z.; Zhao, T.; Xu, W.; Liu, Y.; Zhang, D.; Zhu, D. Synthesis, characterization, and fielde-ffect transistor properties of carbazolenevinylene oligomers: From linear to cyclic architectures. Chem. Eur. J. 2008, 14, 4731–4740. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.K.; Ryu, M.K.; Kim, K.D.; Lee, S.M.; Cho, S.W.; Park, J.W. Tunable electroluminescence from silicon-containing poly(p-phenylenevinylene)-related copolymers with well-defined structures. Macromolecules 1998, 31, 1114–1123. [Google Scholar] [CrossRef]
- Hall, H.K., Jr. Correlation of the Base Strengths of Amines. J. Am. Chem. Soc. 1957, 79, 5441–5444. [Google Scholar] [CrossRef]
- Czerwińska, I.; Juskowiak, B. Photoisomerizable arylstilbazolium ligands recognize parallel and antiparallel structures of G-quadruplexes. Int. J. Biol. Macromol. 2012, 51, 576–582. [Google Scholar] [CrossRef] [PubMed]
- Freyer, M.W.; Buscaglia, R.; Kaplan, K.; Cashman, D.; Hurley, L.H.; Lewis, E.A. Biophysical Studies of the c-MYC NHE III1 Promoter: Model Quadruplex Interactions with a Cationic Porphyrin. Biophys. J. 2007, 92, 2007–2015. [Google Scholar] [CrossRef]
- Slama-Schwok, A.; Rougee, M.; Ibanez, V.; Geacintov, N.E.; Montenay-Garestier, T.; Lehn, J.M.; Hélène, C. Interactions of the dimethyldiazaperopyrenium dication with nucleic acids. 2. Binding to double-stranded polynucleotides. Biochemistry 1989, 28, 3234–3242. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhou, A. Spectroscopic studies on the binding of methylene blue with DNA by means of cyclodextrin supramolecular systems. J. Photochem. Photobiol. A Chem. 2007, 190, 121–127. [Google Scholar] [CrossRef]
- Chan, D.S.-H.; Yang, H.; Kwan, M.H.-T.; Cheng, Z.; Lee, P.; Bai, L.-P.; Jiang, Z.-H.; Wong, C.-Y.; Fong, W.-F.; Leung, C.-H.; et al. Structure-based optimization of FDA-approved drug methylene blue as a c-myc G-quadruplex DNA stabilizer. Biochimie 2011, 93, 1055–1064. [Google Scholar] [CrossRef]
- Bhattacharjee, A.J.; Ahluwalia, K.; Taylor, S.; Jin, O.; Nicoludis, J.M.; Buscaglia, R.; Chaires, J.B.; Kornfilt, D.J.P.; Marquardt, D.G.S.; Yatsunyk, L.A. Induction of G-quadruplex DNA structure by Zn(II) 5,10,15,20-tetrakis (N-methyl-4-pyridyl)porphyrin. Biochimie 2011, 93, 1297–1309. [Google Scholar] [CrossRef]
- Yamashita, T.; Uno, T.; Ishikawa, Y. Stabilization of guanine quadruplex DNA by the binding of porphyrins with cationic side arms. Bioorg. Med. Chem. 2005, 13, 2423–2430. [Google Scholar] [CrossRef]
- Sun, J.; An, Y.; Zhang, L.; Chen, H.-Y.; Han, Y.; Wang, Y.-J.; Mao, Z.-W.; Ji, L.-N. Studies on synthesis, characterization, and G-quadruplex binding of Ru(II) complexes containing two dppz ligands. J. Inorg. Biochem. 2011, 105, 149–154. [Google Scholar] [CrossRef]
- Algar, W.R.; Massey, M.; Krull, U.J. Fluorescence Resonance Energy Transfer and Complex Formation Between Thiazole Orange and Various Dye-DNA Conjugates: Implications in Signaling Nucleic Acid Hybridization. J. Fluorescence 2006, 16, 555–567. [Google Scholar] [CrossRef]
- Crosby, G.A.; Demas, J.N. Measurement of photoluminescence quantum yields. Review. J. Phys. Chem. 1971, 75, 991–1024. [Google Scholar] [CrossRef]
- Paramasivan, S.; Rujan, I.; Bolton, P.H. Circular dichroism of quadruplex DNAs: Applications to structure, cation effects and ligand binding. Methods 2007, 43, 324–331. [Google Scholar] [CrossRef] [PubMed]
- Kypr, J.; Kejnovská, I.; Renciuk, D.; Vorlícková, M. Circular dichroism and conformational polymorphism of DNA. Nucl. Acids Res. 2009, 37, 1713–1725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, E.W.; Tanious, F.; Ismail, M.A.; Reszka, A.P.; Neidle, S.; Boykin, D.W.; Wilson, W.D. Structure-specific recognition of quadruplex DNA by organic cations: Influence of shape, substituents and charge. Biophys. Chem. 2007, 126, 140–153. [Google Scholar] [CrossRef] [PubMed]
- Vorlíčková, M.; Kejnovská, I.; Sagi, J.; Renčiuk, D.; Bednářová, K.; Motlová, J.; Kypr, J. Circular dichroism and guanine quadruplexes. Methods 2012, 57, 64–75. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Tang, Y.; Xiang, J.; Xu, G.; Zhang, Y.; Zhang, H.; Xu, L. Spectroscopic studies of the interaction between quercetin and G-quadruplex DNA. Bioorg. Med. Chem. Lett. 2006, 16, 3586–3589. [Google Scholar] [CrossRef]
- Dash, J.; Shirude, P.S.; Hsu, S.-T.D.; Balasubramanian, S. Diarylethynyl Amides That Recognize the Parallel Conformation of Genomic Promoter DNA G-Quadruplexes. J. Am. Chem. Soc. 2008, 130, 15950–15956. [Google Scholar] [CrossRef] [Green Version]
- Yu, Q.; Liu, Y.; Wang, C.; Sun, D.; Yang, X.; Liu, Y.; Liu, J. Chiral Ruthenium(II) Polypyridyl Complexes: Stabilization of G-Quadruplex DNA, Inhibition of Telomerase Activity and Cellular Uptake. PLoS ONE 2012, 7, e50902. [Google Scholar] [CrossRef] [PubMed]
- Mergny, J.-L.; Phan, A.-T.; Lacroix, L. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998, 435, 74–78. [Google Scholar] [CrossRef] [Green Version]
- Di Antonio, M.; Rodriguez, R.; Balasubramanian, S. Experimental approaches to identify cellular G-quadruplex structures and functions. Methods 2012, 57, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Vy Thi Le, T.; Han, S.; Chae, J.; Park, H.J. G-quadruplex binding ligands: From naturally occurring to rationally designed molecules. Curr. Pharm. Des. 2012, 18, 1948–1972. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, A.; Salvati, E.; Biroccio, A. Methods of studying telomere damage induced by quadruplex-ligand complexes. Methods 2012, 57, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Tataurov, A.V.; You, Y.; Owczarzy, R. Predicting ultraviolet spectrum of single stranded and double stranded deoxyribonucleic acids. Biophys. Chem. 2008, 133, 66–70. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |














| Solvent | ε/104 [M−1∙cm−1] | Abs. λmax [nm] | Em. λmax [nm] |
|---|---|---|---|
| CH2Cl2 | 5.2 ± 0.1 | 501 | 566 |
| EtOH | 5.0 ± 0.1 | 469 | 569 |
| CHCl3 | 5.1 ± 0.1 | 504 | 566 |
| MeOH | 4.9 ± 0.1 | 479 | 569 |
| ACN | 4.8 ± 0.1 | 473 | 570 |
| H2O | 4.3 ± 0.1 | 451 | 565 |
| 10 mM Tris-HCl | 4.3 ± 0.1 | 451 | 566 |
| DMSO | 4.3 ± 0.1 | 468 | 573 |
| 1,4-Dioxane | 1.9 ± 0.1 | 482 | 566 |
| Toluene | 1.9 ± 0.1 | 452 | 565 |
| DNA | Δλmax [nm] a | Hypochromicity [%] b | Hyperchromicity [%] b |
|---|---|---|---|
| G4 22HT Na+ | 33 | 27 | 17 |
| G4 22HT K+ | 33 | 26 | 23 |
| Cations | Benesi–Hildebrand Method, nKb (× 105 M−1) | |
|---|---|---|
| Spectrophotometric Titration | Fluorescence Titration | |
| Na+ | 1.3 ± 0.1 | 0.8 ± 0.1 |
| K+ | 1.5 ± 0.3 | 1.7 ± 0.3 |
| Cations | Tm [°C] | Tm [°C] | ΔTm [°C] e |
|---|---|---|---|
| Na+ | 49.5 a | 54.1 c | 4.5 |
| K+ | 62.0 a | 66.3 c | 4.3 |
| ---- | <14 b | 44.6 d | >30.6 |
| Cytotoxicity (IC50, [μM]) | |||
|---|---|---|---|
| Cell Line | Time Interval | ||
| 24 h | 48 h | 72 h | |
| MCF7 | >18 | 12.5 | 9.5 |
| MDA-MB-231 | >18 | 13.4 | 11.4 |
| MCF-12A | >18 | >18 | >18 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Głuszyńska, A.; Juskowiak, B.; Rubiś, B. Binding Study of the Fluorescent Carbazole Derivative with Human Telomeric G-Quadruplexes. Molecules 2018, 23, 3154. https://doi.org/10.3390/molecules23123154
Głuszyńska A, Juskowiak B, Rubiś B. Binding Study of the Fluorescent Carbazole Derivative with Human Telomeric G-Quadruplexes. Molecules. 2018; 23(12):3154. https://doi.org/10.3390/molecules23123154
Chicago/Turabian StyleGłuszyńska, Agata, Bernard Juskowiak, and Błażej Rubiś. 2018. "Binding Study of the Fluorescent Carbazole Derivative with Human Telomeric G-Quadruplexes" Molecules 23, no. 12: 3154. https://doi.org/10.3390/molecules23123154