Chemical Modification of Sweet Potato β-amylase by Mal-mPEG to Improve Its Enzymatic Characteristics
Abstract
:1. Introduction
2. Results and Discussion
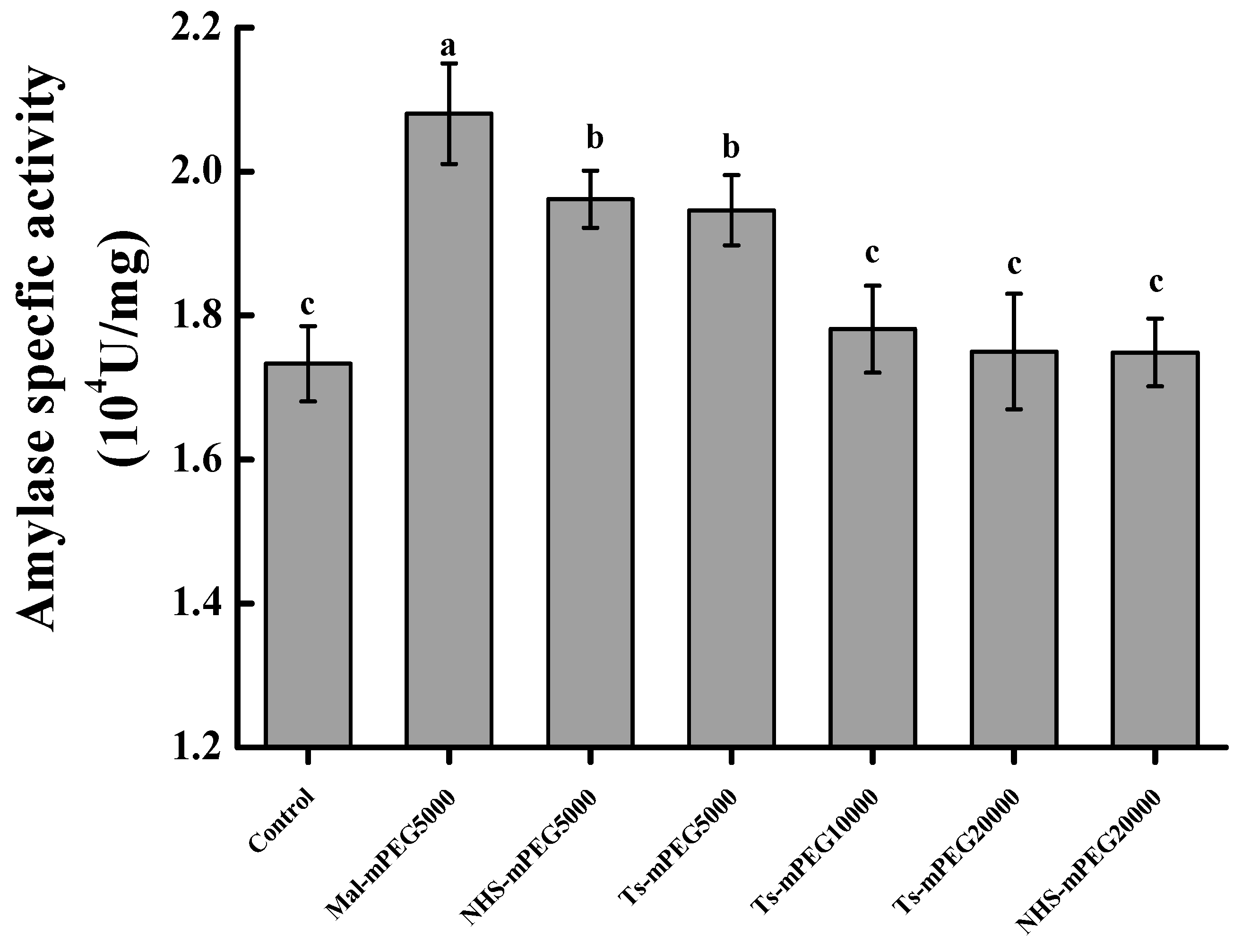
2.1. Screening of Modifiers
2.2. Effect of the Molar Ratio on Modification
2.3. Effect of Temperature on Modification
2.4. Effect of pH on Modification
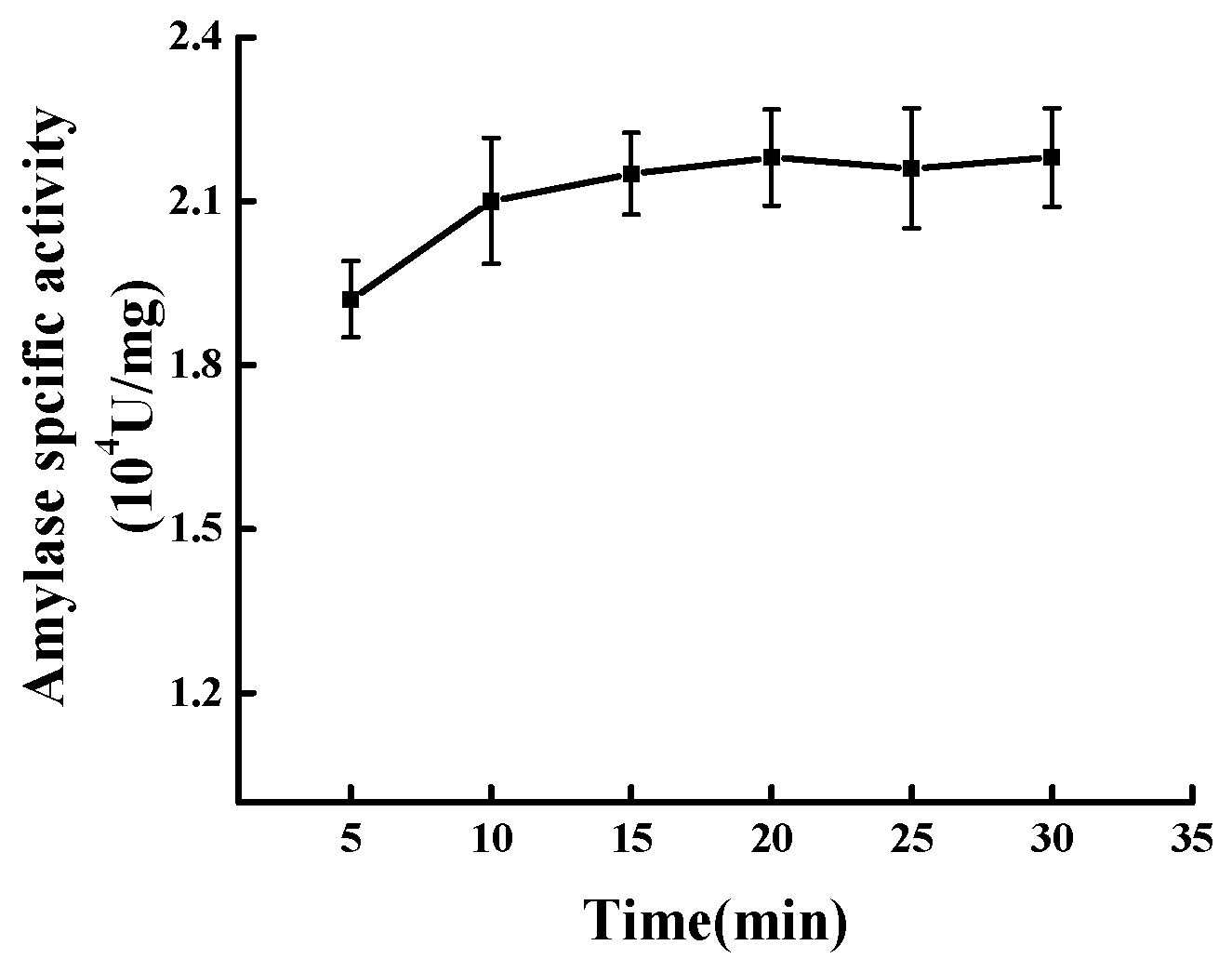
2.5. Effect of Time on Modification
2.6. Optimizing the Modification Procedure
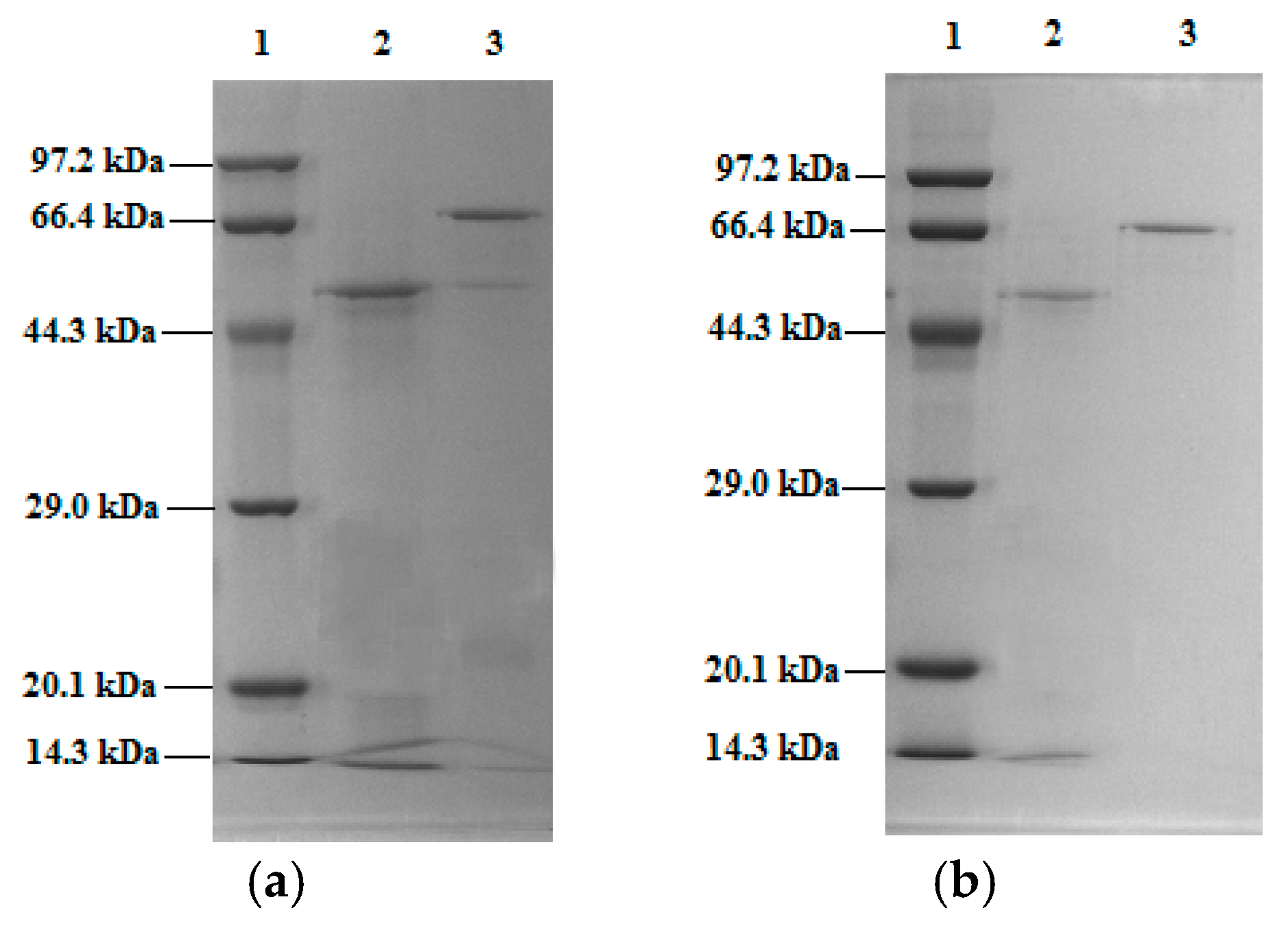
2.7. Separation and Purification of Mal-mPEG5000-SPA
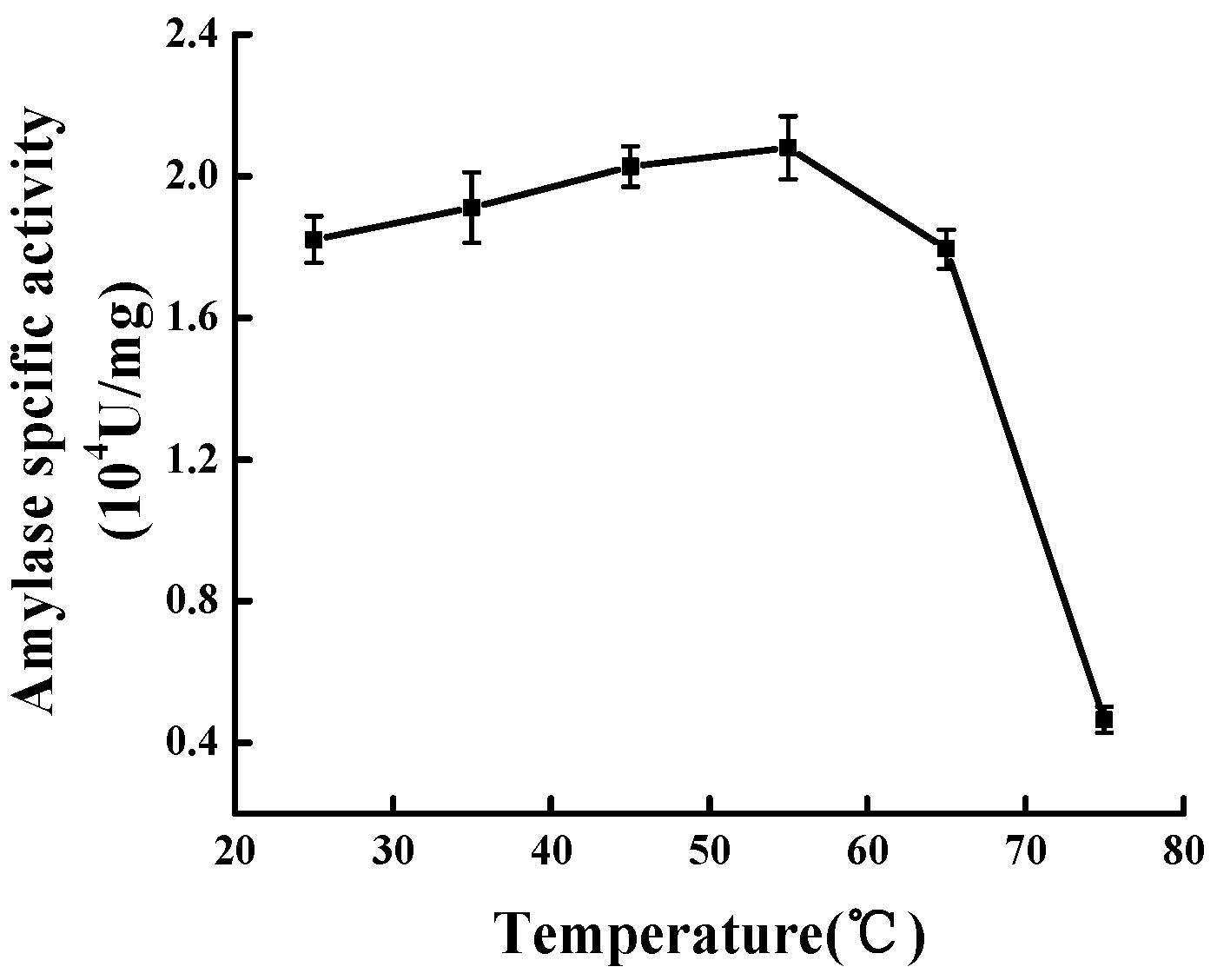
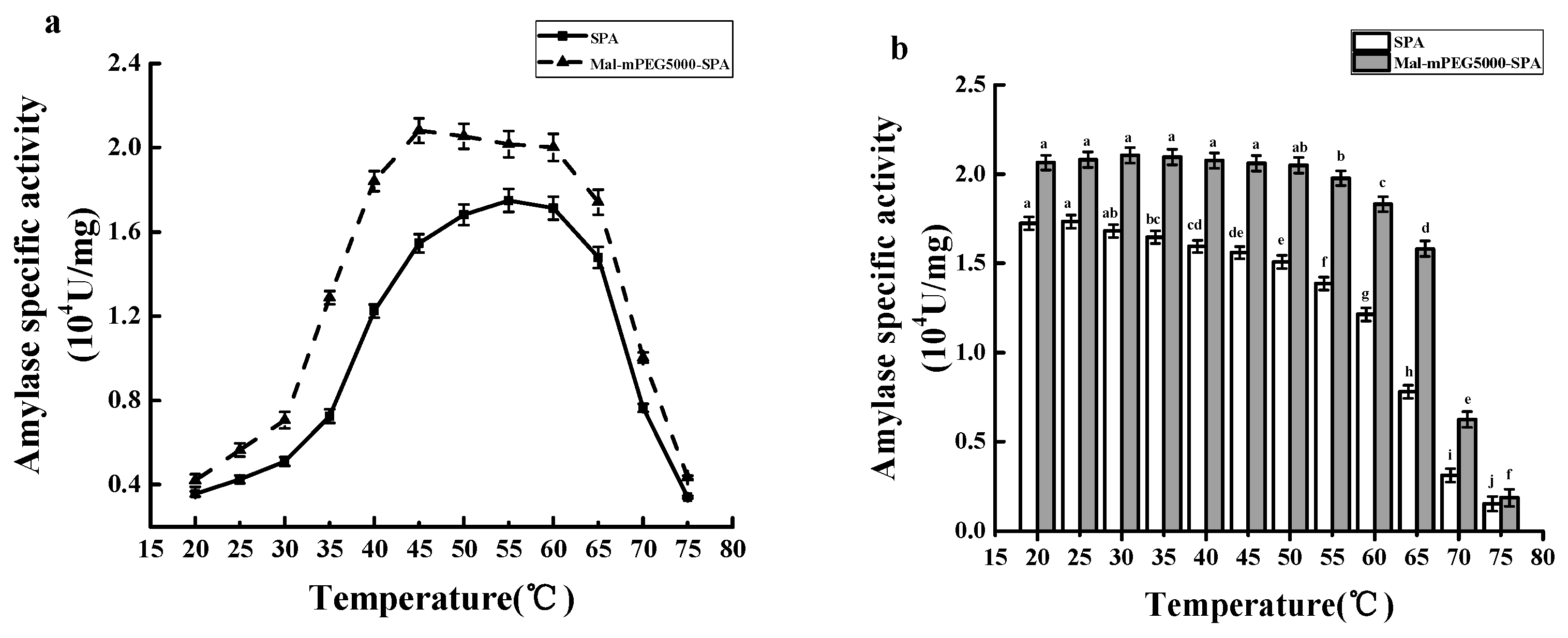
2.8. Optimum Temperature and Thermal Stability of Mal-mPEG5000-SPA and SPA
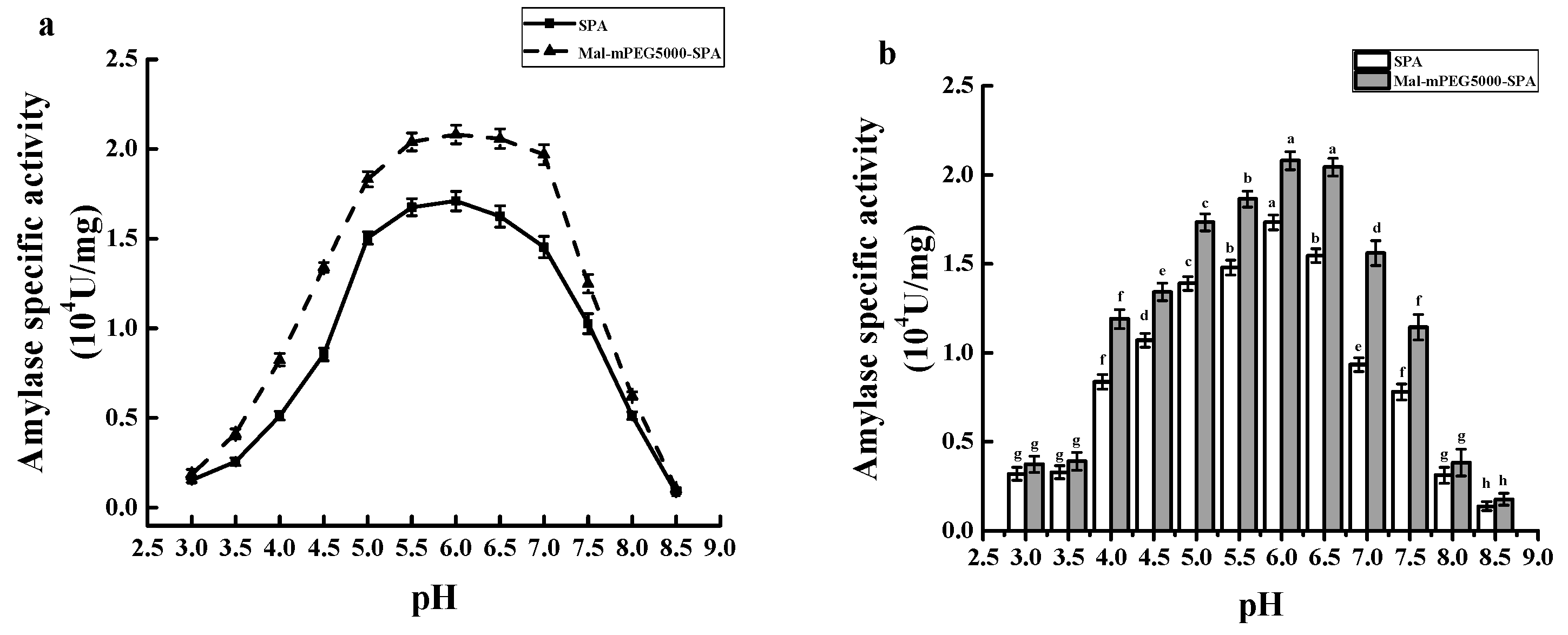
2.9. Optimum pH and pH Stability of Mal-mPEG5000-SPA and SPA
2.10. Kinetic Parameters of Mal-mPEG5000-SPA
2.11. Effect of Metal Ions on the Activity of Mal-mPEG5000-SPA
3. Materials and Methods
3.1. Materials
3.2. Separation and Purification of SPA
3.3. Protein Content and Enzyme Assay
3.4. Screening of Modifiers
3.5. Selection of Relevant Variables and Experimental Design
3.6. Enzyme Characterization
3.7. Effect of Metal Ions on Mal-mPEG5000-SPA and SPA
3.8. Kinetic Constant
3.9. Activation Energy (Ea)
3.10. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Chang, C.T.; Lion, H.Y.; Tang, H.L.; Sung, H.Y. Activation, purification and properties of beta-amylase from sweet potatoes (Ipomoea batatas). Biotechnol. Appl. Biochem. 1996, 126, 120–128. [Google Scholar]
- Dicko, M.H.; Searle-van Leeuwen, M.J.F.; Beldman, G.; Ouedraogo, O.G.; Hilhorst, O.G.; Traore, A.S. Purification and characterization of β-amylase from Curculigo pilosa. Appl. Microbiol. Biotechnol. 1999, 52, 802–805. [Google Scholar] [CrossRef]
- Kaplan, F.; Dong, Y.S.; Guy, C.L. Roles of β-amylase and starch breakdown during temperatures stress. Physiol. Plant. 2010, 126, 120–128. [Google Scholar] [CrossRef]
- Vajravijayan, S.; Pletnev, S.; Mani, N.; Nandhagopal, N.; Gunasekaran, K. Structural insights on starch hydrolysis by plant β-amylase and its evolutionary relationship with bacterial enzymes. Int. J. Biol. Macromol. 2018, 113, 329–337. [Google Scholar] [CrossRef] [PubMed]
- Miao, M.; Li, R.; Huang, C.; Jiang, B.; Zhang, T. Impact of beta-amylase degradation on properties of sugary maize soluble starch particles. Food Chem. 2015, 177, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Mihajlovski, K.R.; Radovanovic, N.R.; Veljovic, D.N.; Siler-Marinkovic, S.S.; Dimitrijevic-Brankovic, S.I. Improved β-amylase production on molasses and sugar beet pulp by a novel strain Paenibacillus chitinolyticus CKS1. Ind. Crops Prod. 2016, 80, 115–122. [Google Scholar] [CrossRef]
- Li, H.S; Oba, K. Major Soluble Proteins of Sweet Potato Roots and Changes in Proteins after Cutting, Infection, or Storage. J. Agric. Chem. Soc. Jpn. 1985, 49, 737–744. [Google Scholar] [Green Version]
- Li, L.; Shang, B.; Hu, L.; Shao, R.; Zhen, Y. Site-specific PEGylation of lidamycin and its antitumor activity. Acta Pharm. Sin. B 2015, 5, 264–269. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, Y.P.; Lin, S.C. Effect of PEGylation on the Activity and Stability of Horseradish Peroxidase and l-N-Carbamoylase in Aqueous Phases. Process Biochem. 2015, 50, 1372–1378. [Google Scholar] [CrossRef]
- Abuchowski, A.; van Es, T.; Palczuk, N.C.; Davis, F.F. Alteration of immunological properties of bovine serum albumin by covalent attachment of polyethylene glycol. J. Biol. Chem. 1977, 252, 3578–3581. [Google Scholar] [PubMed]
- Pfister, D.; Morbidelli, M. Process for protein PEGylation. J. Control. Release Off. J. Control. Release Soc. 2014, 180, 134–149. [Google Scholar] [CrossRef] [PubMed]
- Roberts, M.J.; Bentley, M.D.; Harris, J.M. Chemistry for peptide and protein PEGylation. Adv. Drug Deliv. Rev. 2002, 54, 459–476. [Google Scholar] [CrossRef]
- Dozier, J.K.; Distefano, M.D. Site-Specific PEGylation of Therapeutic Proteins. Int. J. Mol. Sci. 2015, 16, 25831–25864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maiser, B.; Baumgartner, K.; Dismer, F.; Hubbuch, J. Effect of lysozyme solid-phase PEGylation on reaction kinetics and isoform distribution. J. Chromatogr. B 2015, 1002, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Shi, Y.; Zhou, J.; Yang, W.; Bai, L.; Wang, S.; Jin, X.; Niu, Q.; Huang, A.; Wang, D. Structure-based antigenic epitope and PEGylation improve the efficacy of staphylokinase. Microb. Cell Fact 2017, 16, 197. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.G.; Fang, L.M.; Wang, L.; Yin, R.C. Preparation and properties of neutral protease modified with monomethoxypolyethylene glycol. Food Ferment. Ind. 2014, 40, 160–163. [Google Scholar]
- Fang, X.; Wang, X.; Li, G.; Zeng, J.; Li, J.; Liu, J. SS-mPEG chemical modification of recombinant phospholipase C for enhanced thermal stability and catalytic efficiency. Int. J. Biol. Macromol. 2018, 111, 1032–1039. [Google Scholar] [CrossRef] [PubMed]
- Daba, T.; Kojima, K.; Inouye, K. Chemical modification of wheat beta-amylase by trinitrobenzenesulfonic acid, methoxypolyethylene glycol, and glutaraldehyde to improve its thermal stability and activity. Enzyme Microb. Technol. 2013, 53, 420–426. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, S.K.; Basu, T. Study on binding phenomenon of lipase enzyme with tributyrin on the surface of graphene oxide array using surface plasmon resonance. Thin Solid Films 2018, 645, 10–18. [Google Scholar] [CrossRef]
- Liang, X.; Ran, J.; Sun, J.; Wang, T.; Jiao, Z.; He, H.; Zhu, M. Steam-explosion-modified optimization of soluble dietary fiber extraction from apple pomace using response surface methodology. CyTA-J. Food 2018, 16, 1–7. [Google Scholar] [CrossRef]
- Liang, X.; Zhang, W.; Wang, Y.; Sun, J. Purification and characterization of β-amylase from sweet potato (Baizhengshu 2) tuberous roots. Res. J. Biotechnol. 2018, 13, 84–91. [Google Scholar]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Lowry, B.O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, T.J. Protein measurement with Folin phenol reagent. J. Biol. Chem. 1951, 193, 265. [Google Scholar] [PubMed]
- Sagu, S.T.; Nso, E.J.; Homann, T.; Kapseu, C.; Rawel, H.M. Extraction and purification of beta-amylase from stems of Abrus precatorius by three phase partitioning. Food Chem. 2015, 183, 144–153. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Park, S.-H.; Dang, N.D.H.; Duong, H.X.; Duong, T.P.C.; Tran, P.L.; Park, J.-T.; Ni, L.; Park, K.-H. Characterization and thermal inactivation kinetics of highly thermostable ramie leaf β-amylase. Enzyme Microb. Technol. 2017, 101, 17–23. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of SPA and Mal-mPEG5000-β-SPA are available from the authors. |




| Runs | Coded Levels | Response Value | ||
|---|---|---|---|---|
| pH | Molar Ratio | Temperature (°C) | (×104 U/mg) | |
| 1 | −1 | −1 | 0 | 1.7763 |
| 2 | 0 | −1 | −1 | 1.8858 |
| 3 | 0 | 0 | 0 | 2.1592 |
| 4 | 0 | 0 | 0 | 2.1481 |
| 5 | 1 | 0 | −1 | 1.8142 |
| 6 | 0 | 1 | −1 | 1.8667 |
| 7 | 0 | −1 | 1 | 1.8758 |
| 8 | 1 | −1 | 0 | 1.9097 |
| 9 | 0 | 0 | 0 | 2.1323 |
| 10 | −1 | 1 | 0 | 1.8620 |
| 11 | −1 | 0 | 1 | 1.8226 |
| 12 | 0 | 0 | 0 | 2.1597 |
| 13 | 0 | 0 | 0 | 2.1326 |
| 14 | 0 | 1 | 1 | 1.9097 |
| 15 | 1 | 0 | 1 | 1.9097 |
| 16 | 1 | 1 | 0 | 1.8619 |
| 17 | −1 | 0 | −1 | 1.8362 |
| Source | Sum of Squares | df | Mean Square | F-Value | p-Value | Significance |
|---|---|---|---|---|---|---|
| Model | 0.3 | 9 | 0.034 | 145.34 | <0.0001 | ** |
| A | 4.91 × 10−3 | 1 | 4.91 × 10−3 | 21.05 | 0.0025 | ** |
| B | 2.23 × 10−4 | 1 | 2.23 × 10−4 | 0.96 | 0.3606 | |
| C | 1.95 × 10−3 | 1 | 1.95 × 10−3 | 8.36 | 0.0233 | * |
| AB | 4.48 × 10−3 | 1 | 4.48 × 10−3 | 19.23 | 0.0032 | ** |
| AC | 2.98 × 10−3 | 1 | 2.98 × 10−3 | 12.76 | 0.0091 | ** |
| BC | 4.62 × 10−4 | 1 | 4.62 × 10−4 | 1.98 | 0.2022 | |
| A2 | 0.12 | 1 | 0.12 | 507.79 | <0.0001 | ** |
| B2 | 0.067 | 1 | 0.067 | 288.2 | <0.0001 | ** |
| C2 | 0.074 | 1 | 0.074 | 319.52 | <0.0001 | ** |
| Residual | 1.63 × 10−3 | 7 | 2.33 × 10−4 | |||
| Lack of fit | 8.97 × 10−4 | 3 | 2.99 × 10−4 | 1.63 | 0.317 | |
| Pure error | 7.35 × 10−4 | 4 | 1.84 × 10−4 | |||
| Cor total | 0.31 | 16 | ||||
| R2 | 0.996 |
| Substrates | Km (mg/mL) | Vmax (mmol/min/mL) | Ea (kJ/mol) |
|---|---|---|---|
| Sweet potato starch | 1.87 ± 0.032 a | 19.32 ± 0.65 e | 12.67 ± 0.73 a |
| Potato starch | 2.16 ± 0.031 c | 14.62 ± 0.35 c | 19.84 ± 1.03 c |
| Corn starch | 2.56 ± 0.053 e | 9.36 ± 0.27 a | 28.23 ± 1.37 e |
| Soluble starch | 1.96 ± 0.029 b | 16.75 ± 0.68 d | 16.61 ± 0.99 b |
| Amylase | 2.32 ± 0.037 d | 12.85 ± 0.42 b | 22.26 ± 1.09 d |
| Amylopectin | 2.37 ± 0.045 d | 12.53 ± 0.41 b | 22.54 ± 0.98 d |
| Metal Ions | Relative Activity (%) |
|---|---|
| Control | 100 |
| Ca2+ | 102.88 ± 4.32 c |
| Mg2+ | 95.25 ± 4.14 d |
| Cu2+ | 60.95 ± 3.61 f |
| Zn2+ | 111.28 ± 4.82 b |
| Mn2+ | 143.48 ± 6.25 a |
| K+ | 115.49 ± 4.51 b |
| NH4+ | 78.89 ± 4.05 e |
| Hg+ | 5.21 ± 0.63 i |
| Ag+ | 3.28 ± 0.12 i |
| Al3+ | 45.81 ± 3.52 g |
| Fe3+ | 80.85 ± 4.62 e |
| Ba2+ | 59.68 ± 3.92 f |
| Cd2+ | 22.85 ± 2.06 h |
| EDTA | 55.84 ± 3.27 f |
| Factors | Levels | ||
|---|---|---|---|
| −1 | 0 | 1 | |
| Modification pH | 5.0 | 6.0 | 7.0 |
| Molar ratio of SPA to Mal-mPEG5000 | 1:3 | 1:4 | 1:5 |
| Temperature/°C | 45 | 55 | 65 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, X.; Zhang, W.; Ran, J.; Sun, J.; Jiao, L.; Feng, L.; Liu, B. Chemical Modification of Sweet Potato β-amylase by Mal-mPEG to Improve Its Enzymatic Characteristics. Molecules 2018, 23, 2754. https://doi.org/10.3390/molecules23112754
Liang X, Zhang W, Ran J, Sun J, Jiao L, Feng L, Liu B. Chemical Modification of Sweet Potato β-amylase by Mal-mPEG to Improve Its Enzymatic Characteristics. Molecules. 2018; 23(11):2754. https://doi.org/10.3390/molecules23112754
Chicago/Turabian StyleLiang, Xinhong, Wanli Zhang, Junjian Ran, Junliang Sun, Lingxia Jiao, Longfei Feng, and Benguo Liu. 2018. "Chemical Modification of Sweet Potato β-amylase by Mal-mPEG to Improve Its Enzymatic Characteristics" Molecules 23, no. 11: 2754. https://doi.org/10.3390/molecules23112754






