Design, Synthesis, and Evaluation of Amphiphilic Cyclic and Linear Peptides Composed of Hydrophobic and Positively-Charged Amino Acids as Antibacterial Agents
Abstract
:1. Introduction
2. Results and Discussion
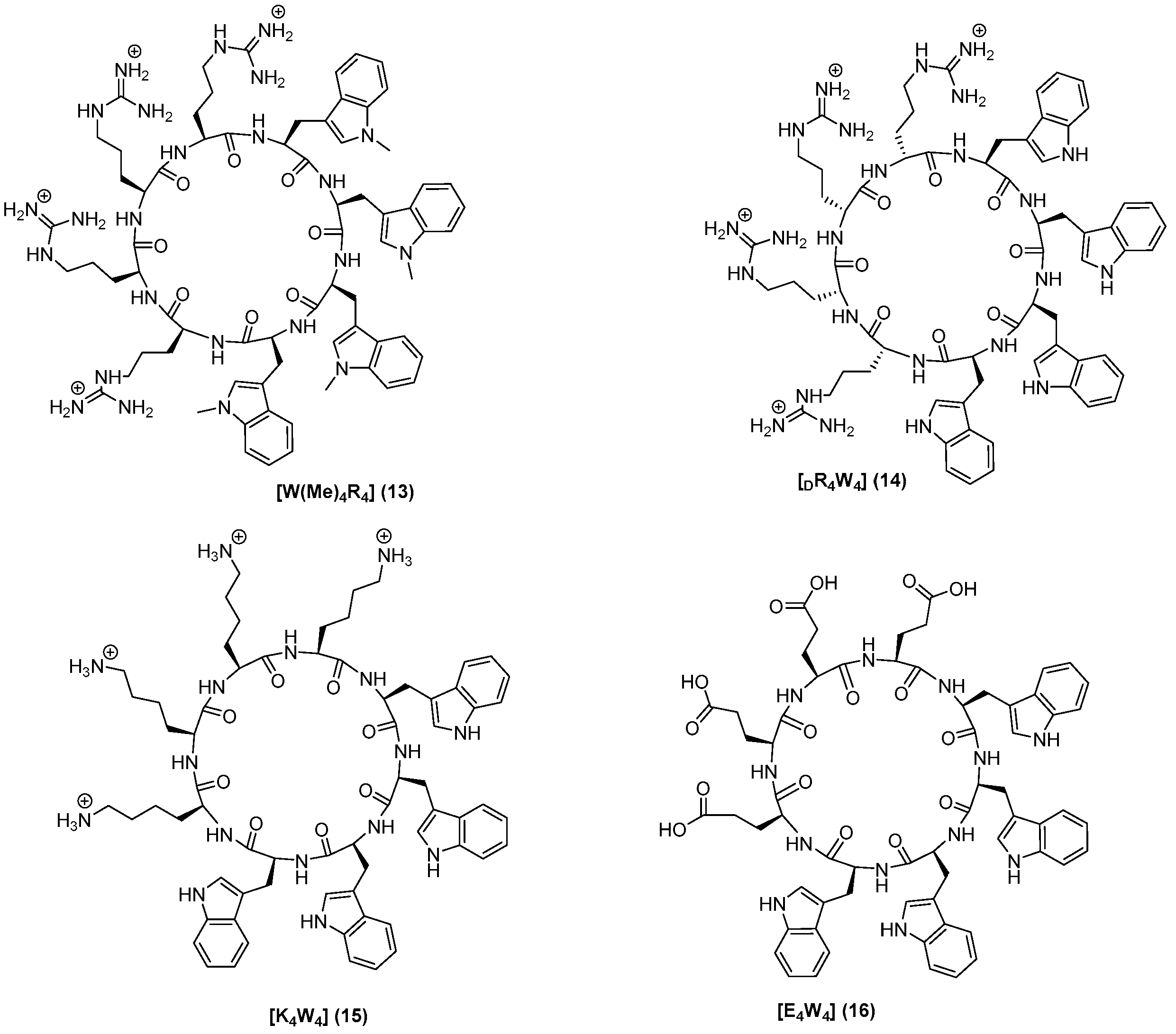
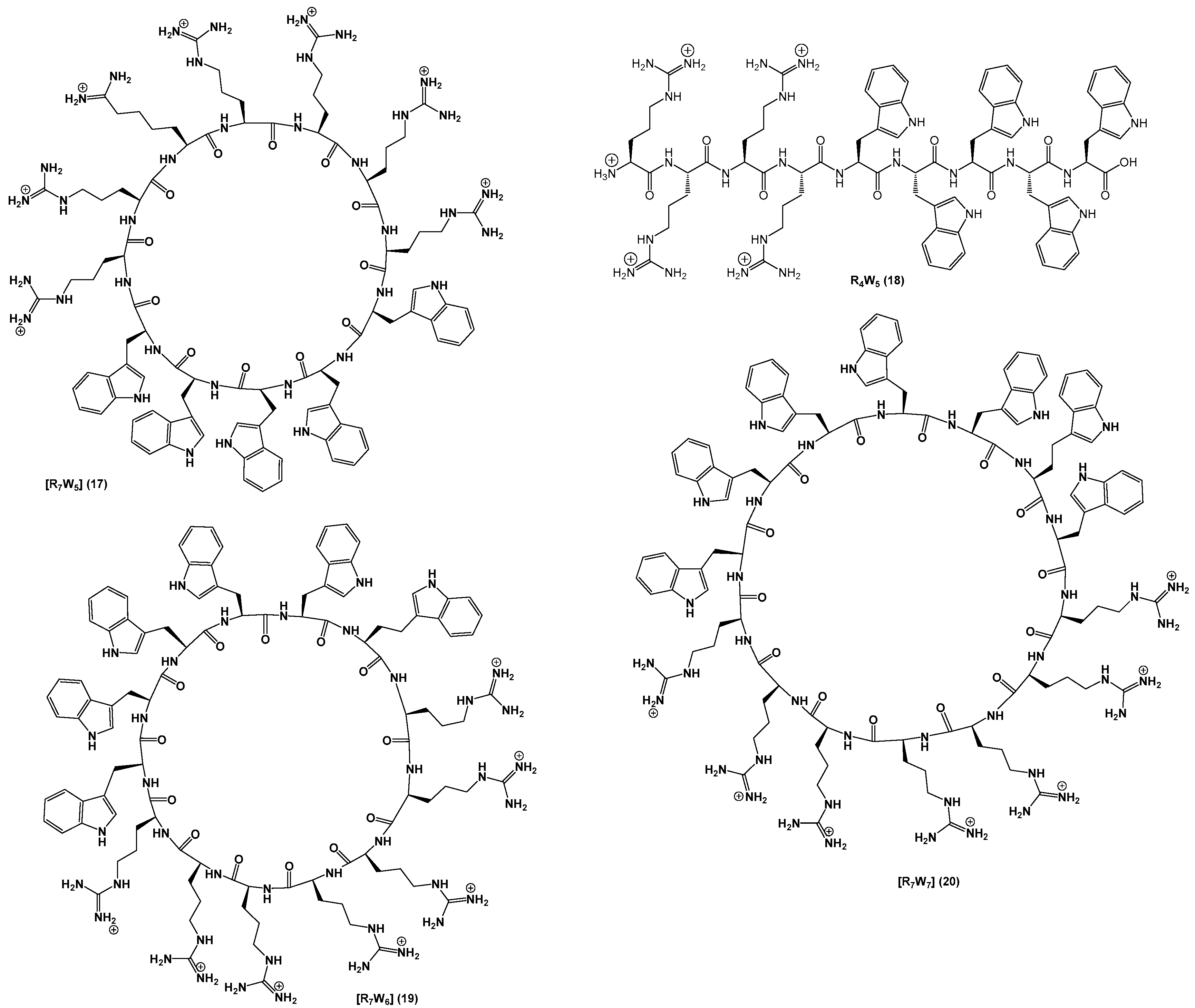
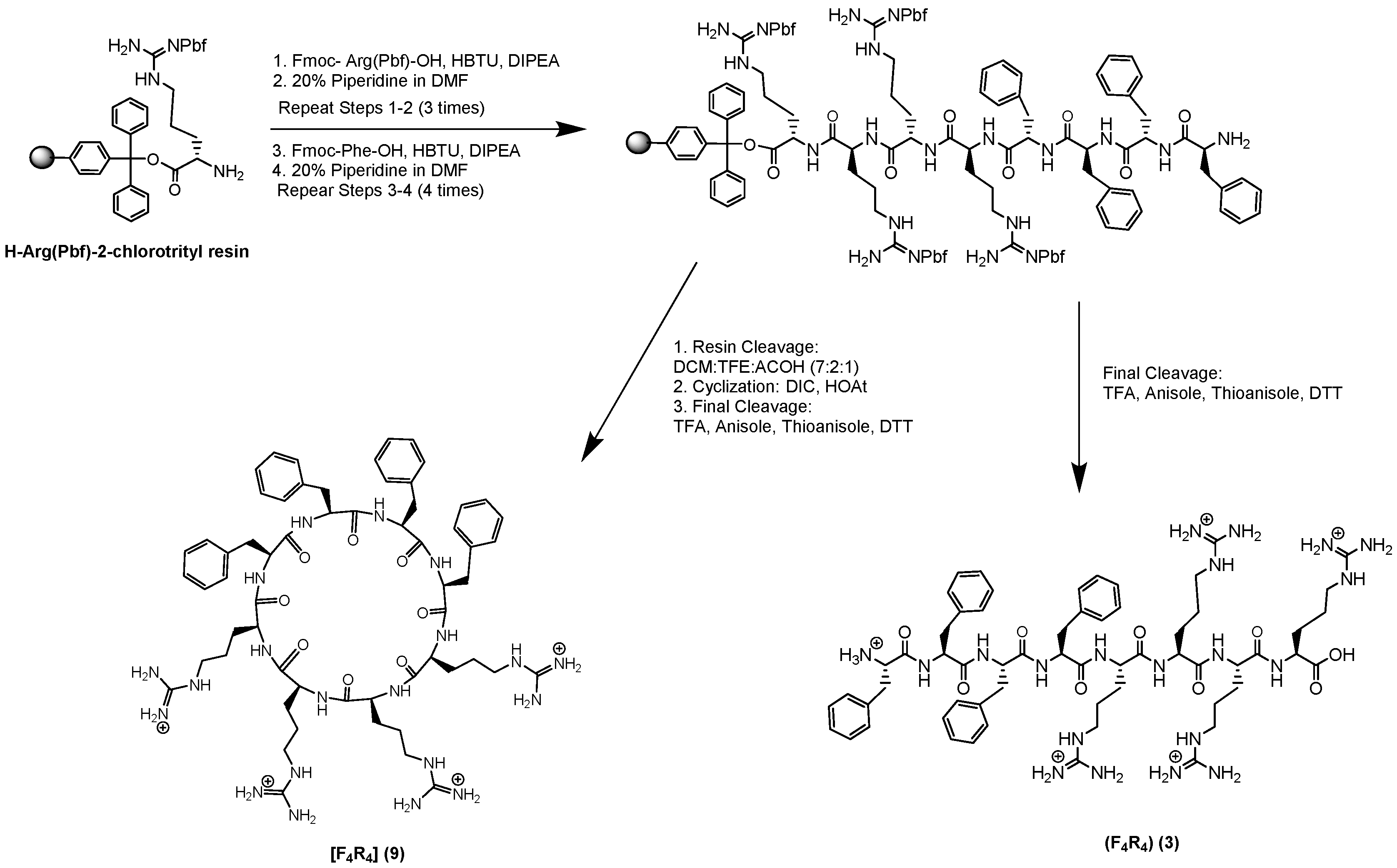
2.1. Chemistry
2.2. Antibacterial Assay
2.3. Cytotoxicity Assay
3. Materials and Methods
3.1. Materials
3.2. Synthesis of Peptides
3.3. Antibacterial Assay
3.4. Cytotoxicity Assay of Peptides
4. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Brown, K.L.; Hancock, R.E. Cationic host defense (antimicrobial) peptides. Curr. Opin. Immunol. 2006, 18, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Wu, H.; Lu, H.; Li, G.; Huang, Q. LAMP: A database linking antimicrobial peptides. PLoS ONE 2013, 8, e66557. [Google Scholar] [CrossRef] [PubMed]
- Radek, K.; Gallo, R. Antimicrobial peptides: Natural effectors of the innate immune system. Semin. Immunopathol. 2007, 29, 27–43. [Google Scholar] [CrossRef] [PubMed]
- Lehrer, R.I.; Ganz, T. Antimicrobial peptides in mammalian and insect host defence. Curr. Opin. Immunol. 1999, 11, 23–27. [Google Scholar] [CrossRef]
- Fjell, C.D.; Hiss, J.A.; Hancock, R.E.; Schneider, G. Designing antimicrobial peptides: Form follows function. Nat. Rev. Drug Discov. 2011, 11, 37–51. [Google Scholar] [CrossRef] [PubMed]
- Costa, F.; Carvalho, I.F.; Montelaro, R.C.; Gomes, P.; Martins, M.C. Covalent immobilization of antimicrobial peptides (AMPs) onto biomaterial surfaces. Acta Biomater. 2011, 7, 1431–1440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wade, J.D.; Lin, F.; Hossain, M.A.; Dawson, R.M. Chemical synthesis and biological evaluation of an antimicrobial peptide gonococcal growth inhibitor. Amino Acids 2012, 43, 2279–2283. [Google Scholar] [CrossRef] [PubMed]
- Azmi, F.; Skwarczynski, M.; Toth, I. Towards the development of synthetic antibiotics: Designs inspired by natural antimicrobial peptides. Curr. Med. Chem. 2016, 23, 4610–4624. [Google Scholar] [CrossRef] [PubMed]
- Matsoukas, J.; Apostolopoulos, V.; Kalbacher, H.; Papini, A.M.; Tselios, T.; Chatzantoni, K.; Biagioli, T.; Lolli, F.; Deraos, S.; Papathanassopoulos, P.; et al. Design and synthesis of a novel potent myelin basic protein epitope 87–99 cyclic analogue: Enhanced stability and biological properties of mimics render them a potentially new class of immunomodulators. J. Med. Chem. 2005, 48, 1470–1480. [Google Scholar] [CrossRef] [PubMed]
- Bogdanowich-Knipp, S.J.; Jois, D.S.; Siahaan, T.J. The effect of conformation on the solution stability of linear vs. cyclic RGD peptides. J. Pept. Res. 1999, 53, 523–529. [Google Scholar] [CrossRef] [PubMed]
- Mandal, D.; Nasrolahi Shirazi, A.; Parang, K. Cell-penetrating homochiral cyclic peptides as nuclear-targeting molecular transporters. Angew. Chem. Int. Ed. Engl. 2011, 50, 9633–9637. [Google Scholar] [CrossRef] [PubMed]
- Hewitt, W.M.; Leung, S.S.; Pye, C.R.; Ponkey, A.R.; Bednarek, M.; Jacobson, M.P.; Lokey, R.S. Cell-permeable cyclic peptides from synthetic libraries inspired by natural products. J. Am. Chem. Soc. 2015, 137, 715–721. [Google Scholar] [CrossRef] [PubMed]
- Schneider, T.; Muller, A.; Miess, H.; Gross, H. Cyclic lipopeptides as antibacterial agents—Potent antibiotic activity mediated by intriguing mode of actions. Int. J. Med. Microbiol. 2014, 304, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Falanga, A.; Nigro, E.; De Biasi, M.G.; Daniele, A.; Morelli, G.; Galdiero, S.; Scudiero, O. Cyclic peptides as novel therapeutic microbicides: Engineering of human defensin mimetics. Molecules 2017, 22, 1217. [Google Scholar] [CrossRef] [PubMed]
- Oh, D.; Sun, J.; Nasrolahi Shirazi, A.; LaPlante, K.L.; Rowley, D.C.; Parang, K. Antibacterial activities of amphiphilic cyclic cell-penetrating peptides against multidrug-resistant pathogens. Mol. Pharm. 2014, 11, 3528–3536. [Google Scholar] [CrossRef] [PubMed]
- Aeschlimann, J.R. The role of multidrug efflux pumps in the antibiotic resistance of Pseudomonas aeruginosa and other gram-negative bacteria. Insights from the Society of Infectious Diseases Pharmacists. Pharmacotherapy 2003, 23, 916–924. [Google Scholar] [CrossRef] [PubMed]
- Riahifard, N.; Tavakoli, K.; Yamaki, J.; Parang, K.; Tiwari, R. Synthesis and evaluation of antimicrobial activity of [R4W4K]-Levofloxacin and [R4W4K]-Levofloxacin-Q conjugates. Molecules 2017, 22, 957. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are available from the authors for a short period of time. |


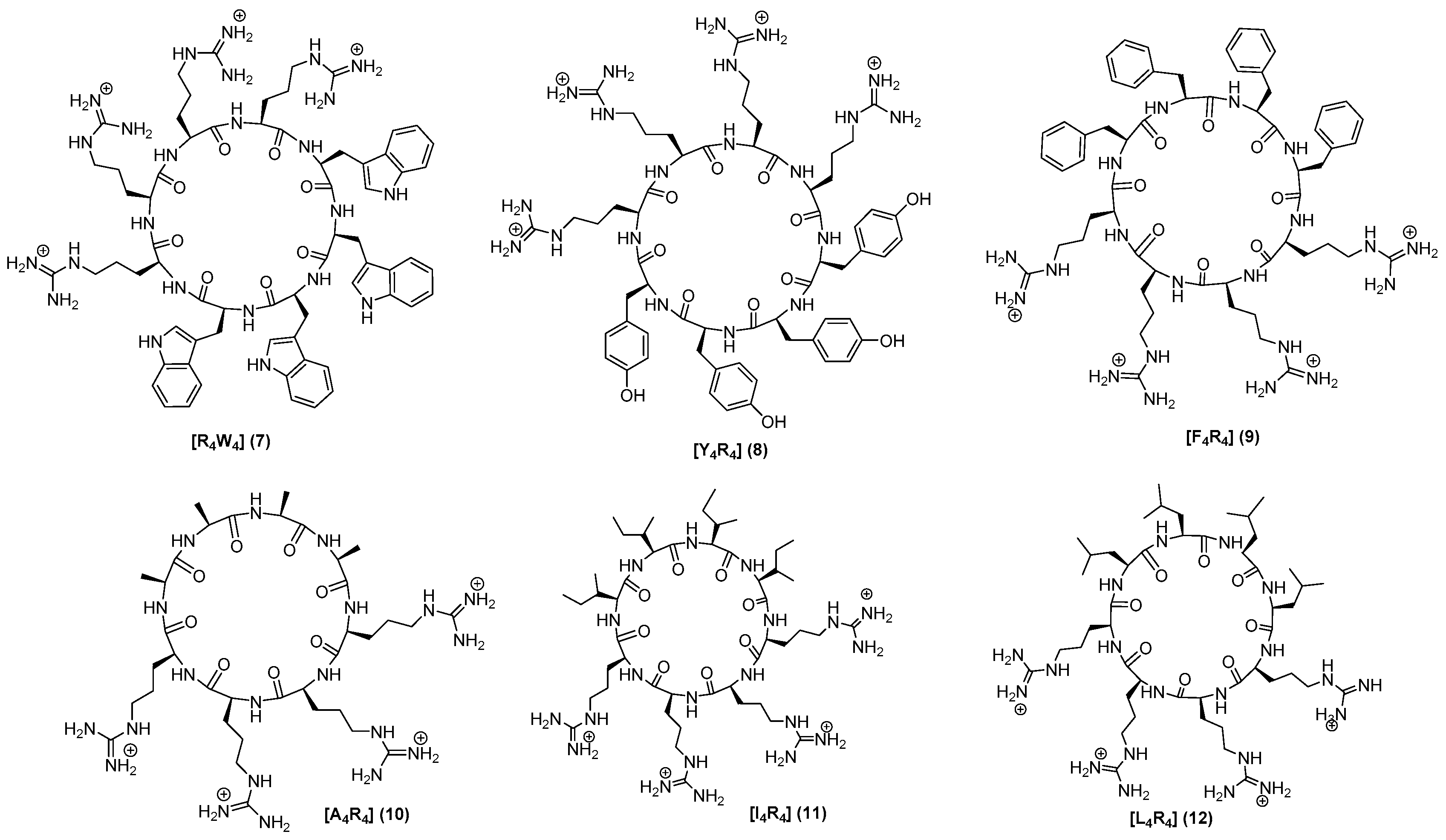

| Compounds | MIC (μg/mL) # MRSA (LAC) | MIC (μg/mL) # E. coli (ATCC 25922) |
|---|---|---|
| R4W4 (1) | 32 | 64 |
| A4R4 (2) | >128 | >128 |
| F4R4 (3) | >128 | >128 |
| L4R4 (4) | >128 | >128 |
| I4R4 (5) | >128 | >128 |
| Y4R4 (6) | >128 | >128 |
| [R4W4] (7) | 4 | 16 |
| [Y4R4] (8) | >128 | >128 |
| [F4R4] (9) | >128 | >128 |
| [A4R4] (10) | >128 | >128 |
| [I4R4] (11) | >128 | >128 |
| [L4R4] (12) | >128 | >128 |
| [W(Me)4R4] (13) | 8 | 16 |
| [DR4W4] (14) | 8 | 16 |
| [K4W4] (15) | 8 | 16 |
| [E4W4] (16) | >128 | >128 |
| [R7W5] (17) | 32 | 64 |
| R4W5 (18)5 | 16 | 32 |
| [R7W6] (19) | 16 | 64 |
| [R7W7] (20) | 8 | 32 |
| Vancomycin Meropenem | 1 ND | ND 0.25 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Riahifard, N.; Mozaffari, S.; Aldakhil, T.; Nunez, F.; Alshammari, Q.; Alshammari, S.; Yamaki, J.; Parang, K.; Tiwari, R.K. Design, Synthesis, and Evaluation of Amphiphilic Cyclic and Linear Peptides Composed of Hydrophobic and Positively-Charged Amino Acids as Antibacterial Agents. Molecules 2018, 23, 2722. https://doi.org/10.3390/molecules23102722
Riahifard N, Mozaffari S, Aldakhil T, Nunez F, Alshammari Q, Alshammari S, Yamaki J, Parang K, Tiwari RK. Design, Synthesis, and Evaluation of Amphiphilic Cyclic and Linear Peptides Composed of Hydrophobic and Positively-Charged Amino Acids as Antibacterial Agents. Molecules. 2018; 23(10):2722. https://doi.org/10.3390/molecules23102722
Chicago/Turabian StyleRiahifard, Neda, Saghar Mozaffari, Taibah Aldakhil, Francisco Nunez, Qamar Alshammari, Saud Alshammari, Jason Yamaki, Keykavous Parang, and Rakesh Kumar Tiwari. 2018. "Design, Synthesis, and Evaluation of Amphiphilic Cyclic and Linear Peptides Composed of Hydrophobic and Positively-Charged Amino Acids as Antibacterial Agents" Molecules 23, no. 10: 2722. https://doi.org/10.3390/molecules23102722





