Increasing Antiradical Activity of Polyphenols from Lotus Seed Epicarp by Probiotic Bacteria Bioconversion
Abstract
:1. Introduction
2. Results and Discussion
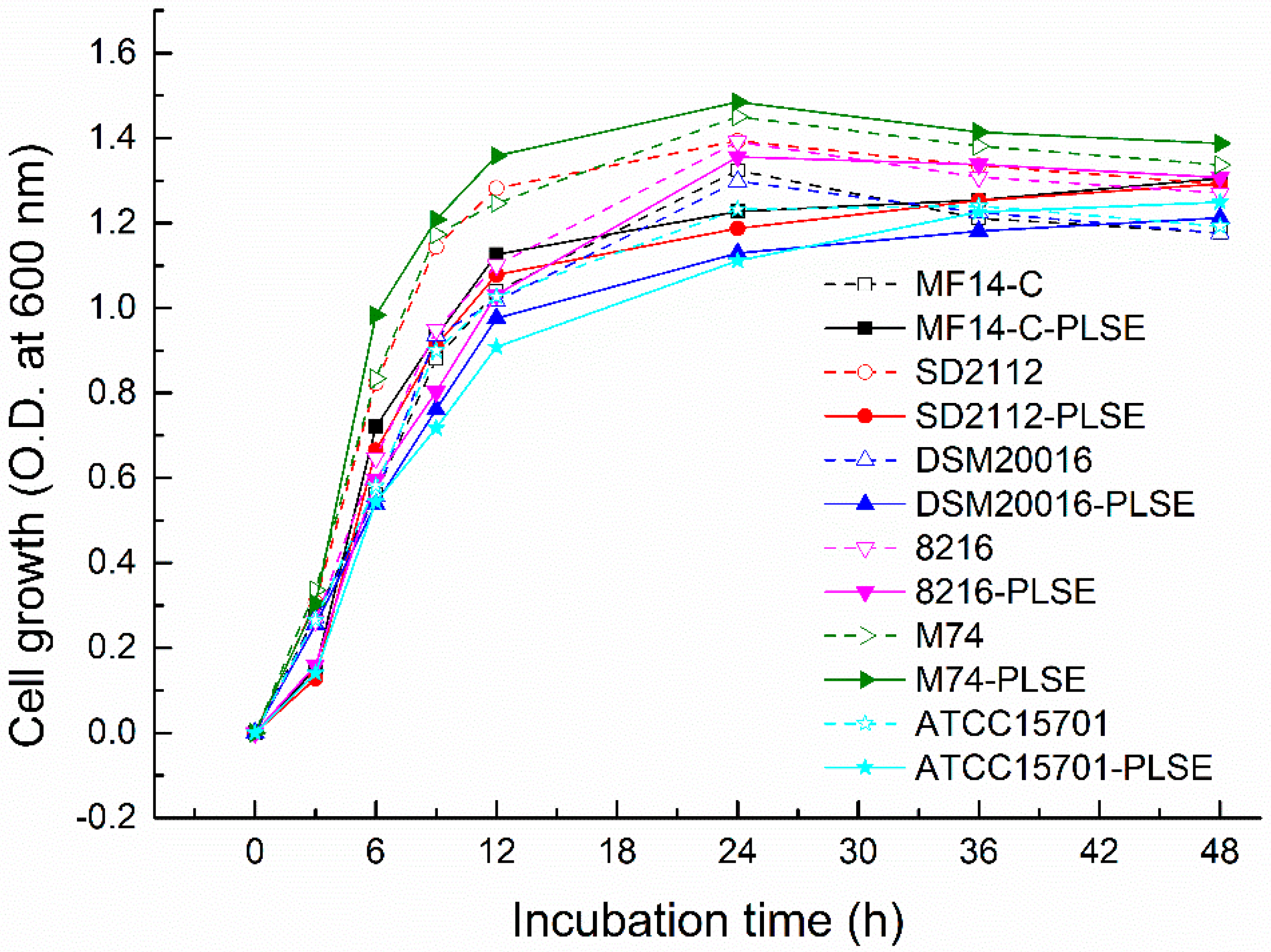
2.1. Effect of PLSE on the Growth of Probiotic Bacteria
2.2. ABTS+ Radical Scavenging Ability of Metabolized PLSE
2.3. DPPH Radical Scavenging Ability of Metabolized PLSE
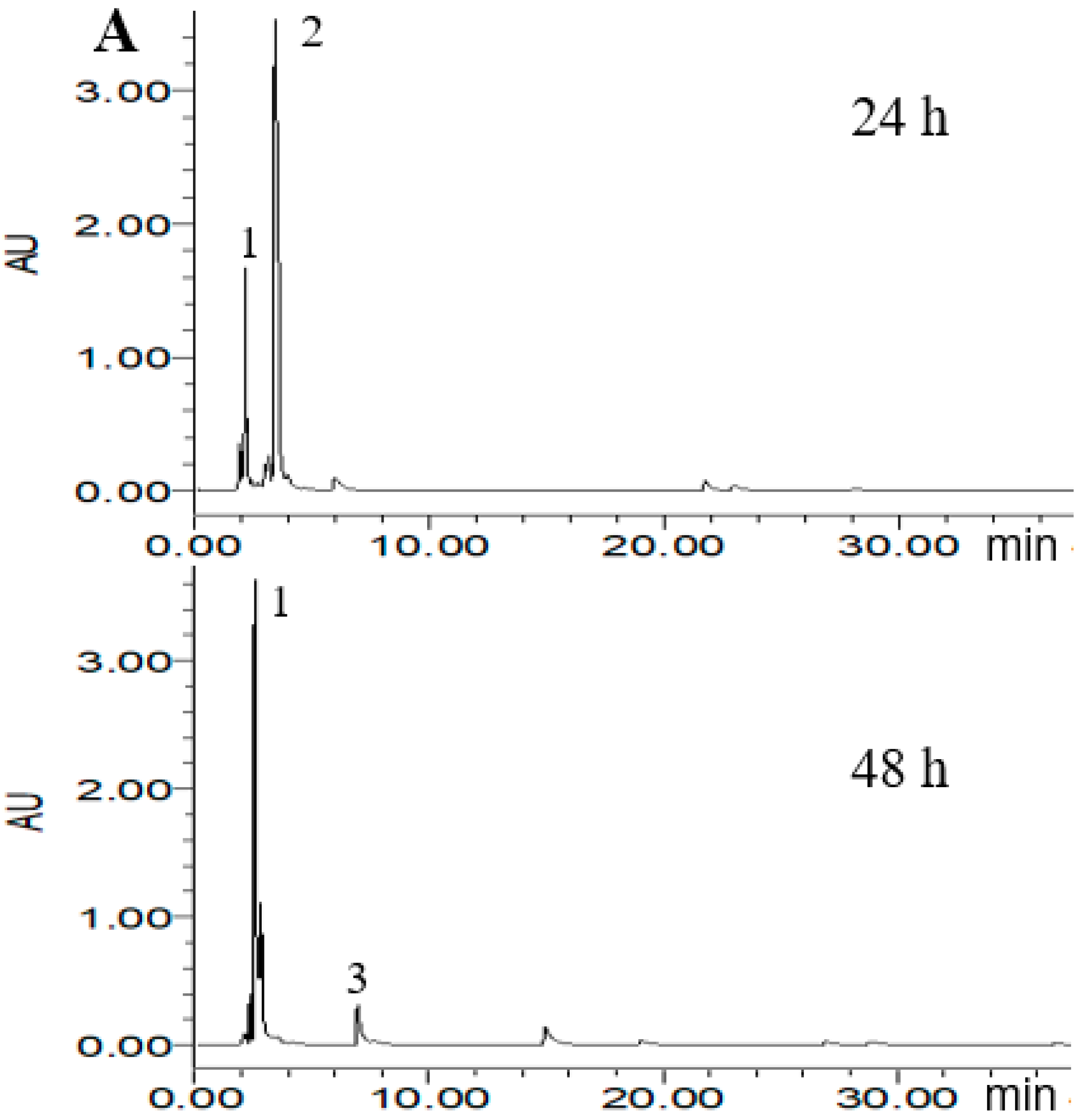
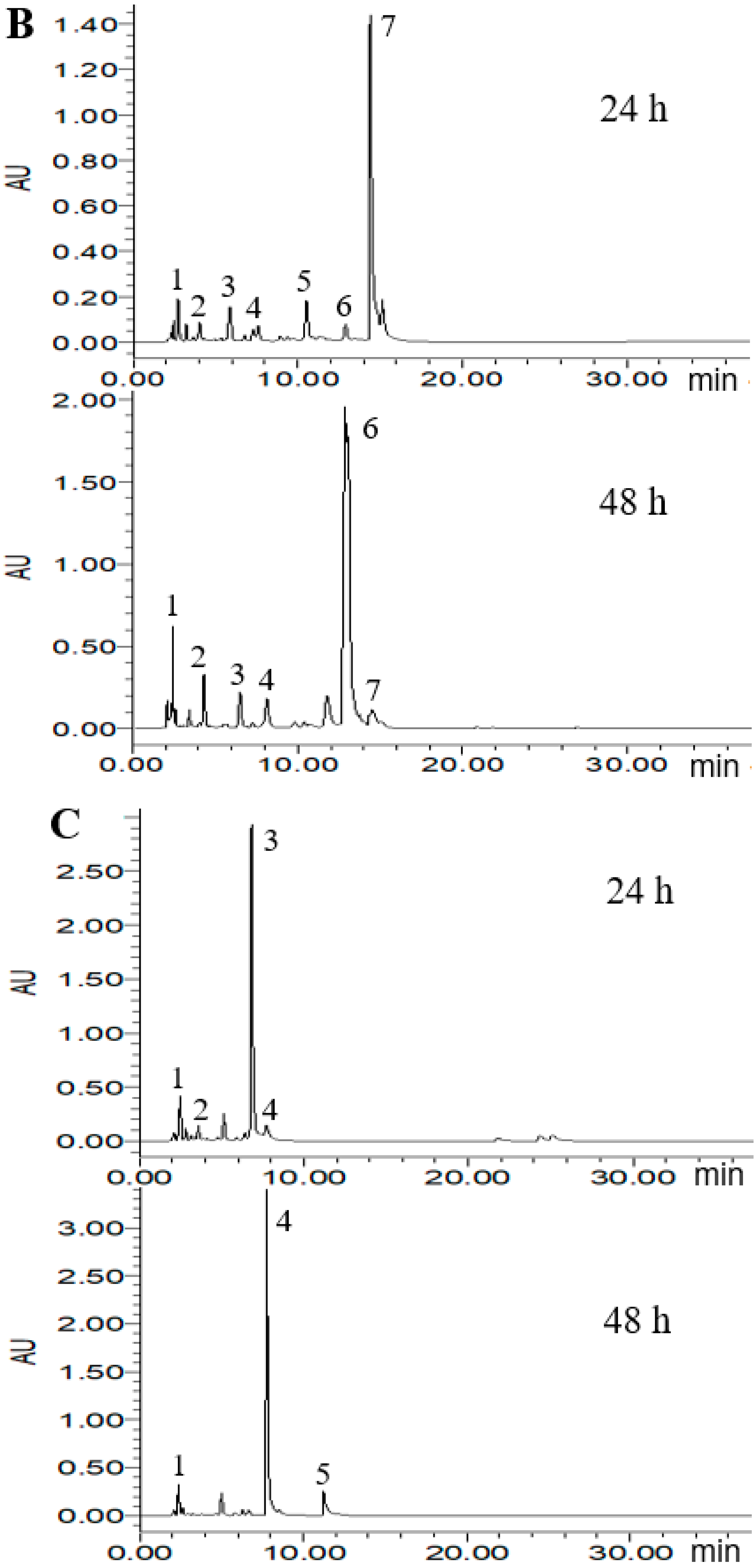
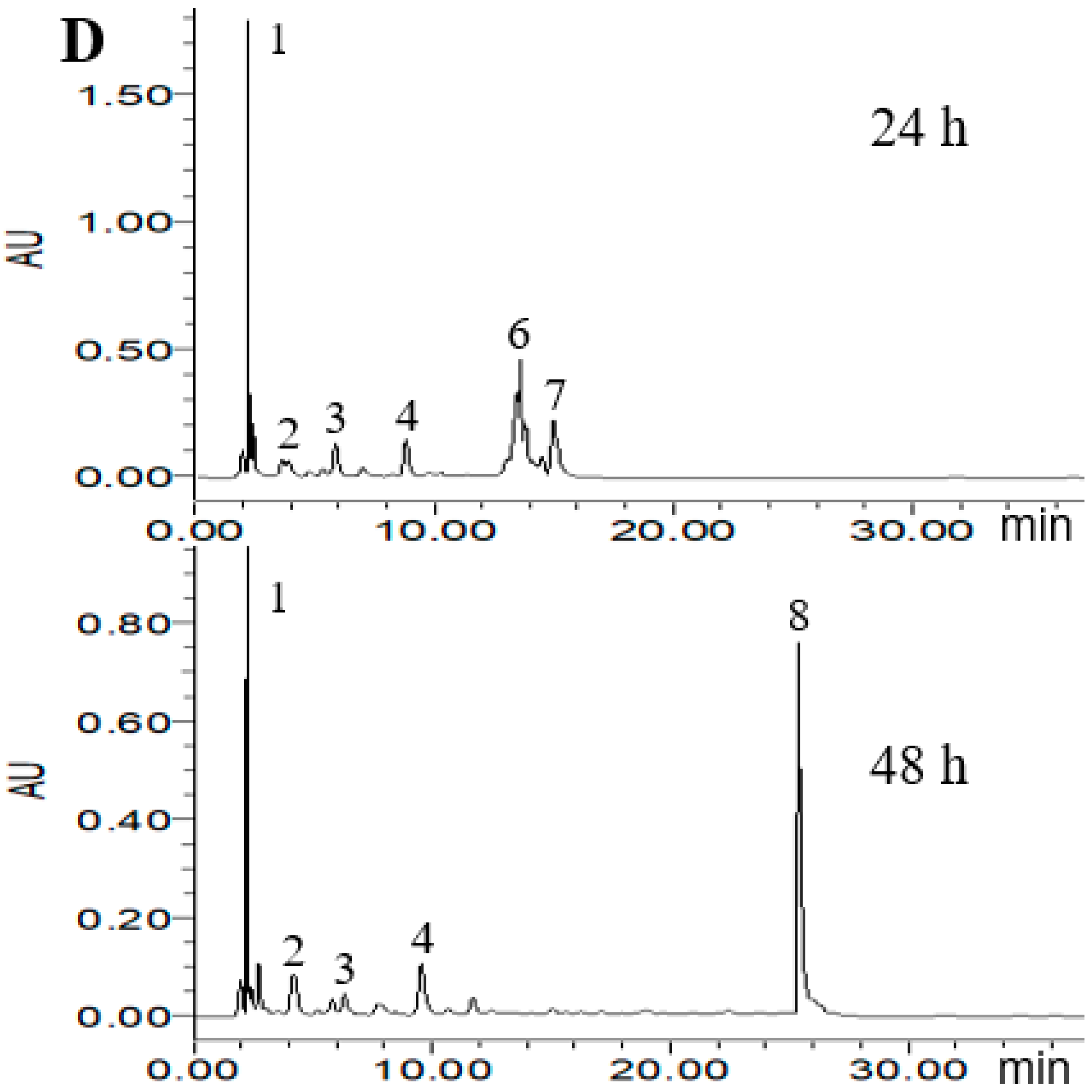
2.4. HPLC Analysis of Metabolized PLSE
3. Materials and Methods
3.1. Probiotic Bacteria and Growth Conditions
3.2. Extraction of PLSE
3.3. Bacterial Activity on PLSE
3.4. ABTS+ Radical Scavenging Activity
3.5. DPPH Radical Scavenging Activity
3.6. HPLC analysis
3.7. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wu, S.; Sun, C.; Cao, X.; Zhou, H.; Zhang, H.; Pan, Y. Preparative counter-current chromatography isolation of liensinine and its analogues from embryo of the seed of Nelumbo nucifera GAERTN. using upright coil planet centrifuge with four multilayer coils connected in series. J. Chromatogr. A 2004, 1041, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.J.; Shin, H.S. Antioxidative effect of lotus seed and seedpod extracts. Food Sci. Biotechnol. 2012, 21, 1761–1766. [Google Scholar] [CrossRef]
- Liu, C.P.; Tsai, W.J.; Lin, Y.L.; Liao, J.F.; Chen, C.F.; Kuo, Y.C. The extracts from Nelumbo Nucifera suppress cell cycle progression, cytokine genes expression, and cell proliferation in human peripheral blood mononuclear cells. Life Sci. 2004, 75, 699–716. [Google Scholar] [CrossRef] [PubMed]
- Kashiwada, Y.; Aoshima, A.; Ikeshiro, Y.; Chen, Y.P.; Furukawa, H.; Itoigawa, M.; Fujioka, T.; Mihashi, K.; Cosentino, L.M.; Morris-Natschke, S.L.; et al. Anti-HIV benzylisoquinoline alkaloids and flavonoids from the leaves of Nelumbo nucifera, and structure-activity correlations with related alkaloids. Bioorg. Med. Chem. 2005, 13, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Rai, S.; Wahile, A.; Mukherjee, K.; Saha, B.P.; Mukherjee, P.K. Antioxidant activity of Nelumbo nucifera (sacred lotus) seeds. J. Ethnopharmacol. 2006, 104, 322–327. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.Z.; Wei, X.L.; Gao, F.F.; Wang, L.S.; Zhang, H.J.; Xu, Y.J.; Li, C.H.; Ge, Y.X.; Zhang, J.J.; Zhang, J. Simultaneous analysis of anthocyanins and flavonols in petals of lotus (Nelumbo) cultivars by high-performance liquid chromatography-photodiode array detection/electrospray ionization mass spectrometry. J. Chromatogr. A 2009, 1216, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Wang, D.; Duan, W.; Du, J.; Wang, X. Preparative isolation and purification of four flavonoids from the petals of Nelumbo nucifera by high-speed counter-current chromatography. Phytochem. Anal. 2010, 21, 268–272. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, P.K.; Saha, K.; Das, J.; Pal, M.; Saha, B.P. Studies on the anti-inflammatory activity of rhizomes of nelumbo nucifera. Planta Med. 1997, 63, 367–369. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Chen, S.; Yin, X.; Wang, K.; Liu, Y.; Li, S.; Yang, P. Systematic qualitative and quantitative assessment of anthocyanins, flavones and flavonols in the petals of 108 lotus (Nelumbo nucifera) cultivars. Food Chem. 2013, 139, 307–312. [Google Scholar] [CrossRef] [PubMed]
- Li, S.S.; Wu, J.; Chen, L.G.; Du, H.; Xu, Y.J.; Wang, L.J.; Zhang, H.J.; Zheng, X.C.; Wang, L.S. Biogenesis of c-glycosyl flavones and profiling of flavonoid glycosides in lotus (Nelumbo nucifera). PLoS ONE 2014, 9, e108860. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.L.; Wang, L.; Wang, E.J.; Zhang, G.L.; Chen, B.; Wang, M.K.; Li, F. Flavonoid glycosides and alkaloids from the embryos of Nelumbo nucifera, seeds and their antioxidant activity. Fitoterapia 2018, 125, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Kredy, H.M.; Huang, D.; Xie, B.; He, H.; Yang, E.; Tian, B.; Xiao, D. Flavonols of lotus (Nelumbo nucifera, Gaertn.) seed epicarp and their antioxidant potential. Eur. Food Res. Technol. 2010, 231, 387–394. [Google Scholar] [CrossRef]
- Chen, S.; Fang, L.; Xi, H.; Guan, L.; Fang, J.; Liu, Y.; Wu, B.; Li, S. Simultaneous qualitative assessment and quantitative analysis of flavonoids in various tissues of lotus (Nelumbo nucifera) using high performance liquid chromatography coupled with triple quad mass spectrometry. Anal. Chim. Acta 2012, 724, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ma, S.S.; Ibrahim, S.A.; Li, E.H.; Yang, H.; Huang, W. Identification and antioxidant properties of polyphenols in lotus seed epicarp at different ripening stages. Food Chem. 2015, 185, 159–164. [Google Scholar] [CrossRef] [PubMed]
- Selma, M.V.; Espín, J.C.; Tomás-Barberán, F.A. Interaction between phenolics and gut microbiota, role in human health. J. Agric. Food Chem. 2009, 57, 6485–6501. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, H.; Curiel, J.A.; Landete, J.M.; de las Rivas, B.; de Felipe, F.L.; Gómez-Cordovés, C.; Mancheño, J.M.; Muñoz, R. Food phenolics and lactic acid bacteria. Int. J. Food Microbiol. 2009, 132, 79–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tabasco, R.; Sánchez-Patán, F.; Monagas, M.; Bartolomé, B.; Moreno-Arribas, M.V.; Peláez, C.; Requena, T. Effect of grape polyphenols on lactic acid bacteria and bifidobacteria growth, Resistance and metabolism. Food Microbiol. 2011, 28, 1345–1352. [Google Scholar] [CrossRef] [PubMed]
- Jakesevic, M.; Aaby, K.; Borge, G.-I.A.; Jeppsson, B.; Ahrné, S.; Molin, G. Antioxidative protection of dietary bilberry, chokeberry and Lactobacillus plantarum HEAL19 in mice subjected to intestinal oxidative stress by ischemia-reperfusion. BMC Complement. Altern. Med. 2011, 11, 8. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Chen, L.; Yang, T.; Wu, Q.; Lv, Z.; Xie, B.; Sun, Z. Increasing antioxidant activity of procyanidin extracts from the pericarp of Litchi chinensis processing waste by two probiotic bacteria bioconversions. J. Agric. Food Chem. 2013, 61, 2506–2512. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Cheng, G.; Han, T.; Yang, H.; Ibrahim, S.A.; Huang, W. Microbial transformation of tectoridin from Pueraria flos by Lactobacillus and bifidobacteria. Food Chem. 2012, 131, 149–154. [Google Scholar] [CrossRef]
- Hervert-Hernández, D.; Pintado, C.; Rotger, R.; Goñi, I. Stimulatory role of grape pomace polyphenols on Lactobacillus acidophilus growth. Int. J. Food Microbiol. 2009, 136, 119–122. [Google Scholar] [CrossRef] [PubMed]
- Pyo, Y.H.; Lee, T.C.; Lee, Y.C. Enrichment of bioactive isoflavones in soymilk fermented with β-glucosidase-producing lactic acid bacteria. Food Res. Int. 2005, 38, 551–559. [Google Scholar] [CrossRef]
- Han, T.; Cheng, G.; Liu, Y.; Yang, H.; Hu, Y.T.; Huang, W. In vitro evaluation of tectoridin, tectorigenin and tectorigenin sodium sulfonate on antioxidant properties. Food Chem. Toxicol. 2012, 50, 409–414. [Google Scholar] [CrossRef] [PubMed]
- Rúa, J.; López-Rodríguez, I.; Sanz, J.; García-Fernández, M.C.; del Valle, M.P.; García-Armesto, M.R. Improving functional properties of “Piel de Sapo” melon juice by addition of a Lippia citriodora, natural extract and probiotic-type lactic acid bacteria. LWT-Food Sci. Technol. 2018, 96, 75–81. [Google Scholar] [CrossRef]
- Vamanu, E.; Gatea, F.; Sârbu, I. In vitro ecological response of the human gut microbiome to bioactive extracts from edible wild mushrooms. Molecules 2018, 23, 2128. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Díaz, I.; Fernández-Navarro, T.; Salazar, N.; Bartolomé, B.; Moreno-Arribas, M.V.; de Andres-Galiana, E.J.; Fernández-Martínez, J.L.; de los Reyes-Gavilán, C.G.; Gueimonde, M.; González, S. Adherence to a mediterranean diet influences the fecal metabolic profile of microbial-derived phenolics in a Spanish cohort of middle-age and older people. J. Agric. Food Chem. 2017, 65, 586–595. [Google Scholar] [CrossRef] [PubMed]
- Areea, T.; Jongrungruangchok, S. Crystallographic evidence for β-cyclodextrin inclusion complexation facilitating the improvement of antioxidant activity of tea (+)-catechin and (−)-epicatechin. Carbohydr. Polym. 2016, 140, 362–373. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Liu, D.; Sun, J.; Yang, X.; Fu, M.; Guo, Y. Simultaneous separation and purification of chlorogenic acid, epicatechin, hyperoside and phlorizin from thinned young Qinguan apples by successive use of polyethylene and polyamide resins. Food Chem. 2017, 230, 362–371. [Google Scholar] [CrossRef] [PubMed]
- Re, R.; Pellegrini, N.; Proteggente, A.; Pannala, A.; Yang, M.; Rice-Evans, C. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Radic. Biol. Med. 1999, 26, 1231–1237. [Google Scholar] [CrossRef]
- Braca, A.; De Tommasi, N.; Di Bari, L.; Pizza, C.; Politi, M.; Morelli, I. Antioxidant Principles from Bauhinia tarapotensis. J. Nat. Prod. 2001, 64, 892–895. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the PLSE are available from the authors. |


© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Hui, X.; Ibrahim, S.A.; Huang, W. Increasing Antiradical Activity of Polyphenols from Lotus Seed Epicarp by Probiotic Bacteria Bioconversion. Molecules 2018, 23, 2667. https://doi.org/10.3390/molecules23102667
Liu Y, Hui X, Ibrahim SA, Huang W. Increasing Antiradical Activity of Polyphenols from Lotus Seed Epicarp by Probiotic Bacteria Bioconversion. Molecules. 2018; 23(10):2667. https://doi.org/10.3390/molecules23102667
Chicago/Turabian StyleLiu, Ying, Xuekuan Hui, Salam A. Ibrahim, and Wen Huang. 2018. "Increasing Antiradical Activity of Polyphenols from Lotus Seed Epicarp by Probiotic Bacteria Bioconversion" Molecules 23, no. 10: 2667. https://doi.org/10.3390/molecules23102667





