Flavones as Quorum Sensing Inhibitors Identified by a Newly Optimized Screening Platform Using Chromobacterium violaceum as Reporter Bacteria
Abstract
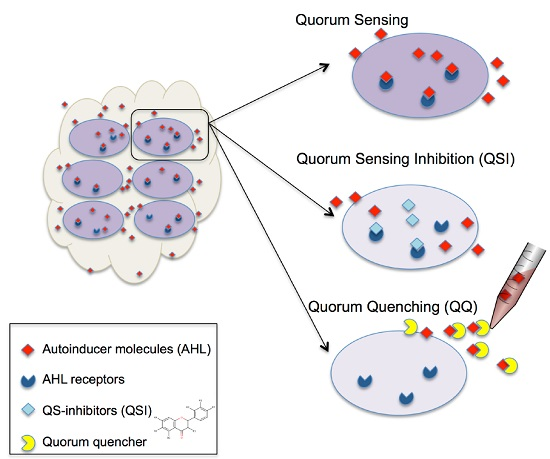
:1. Introduction
2. Results and Discussion
2.1. Optimization of Growing Conditions and Violacein Production
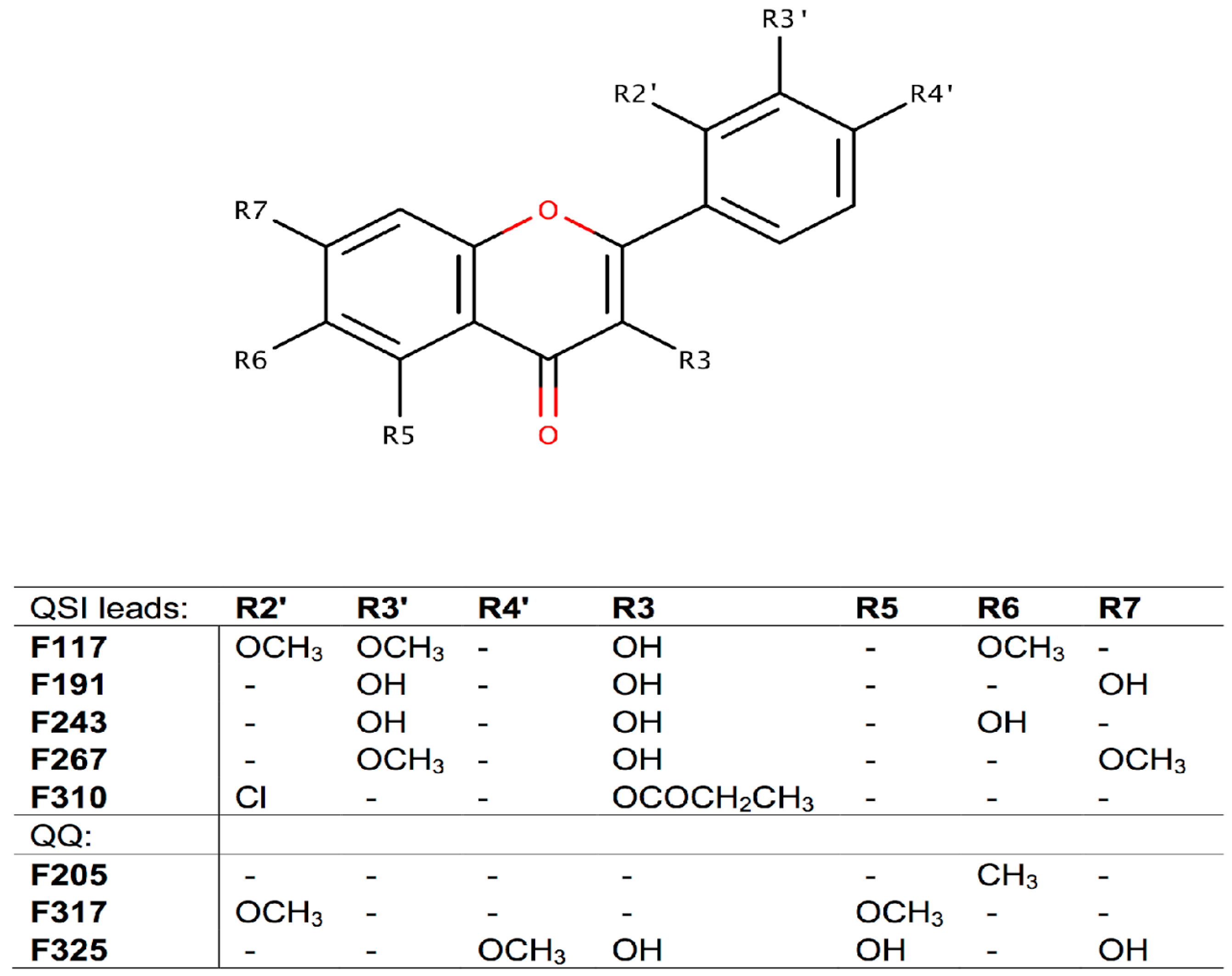
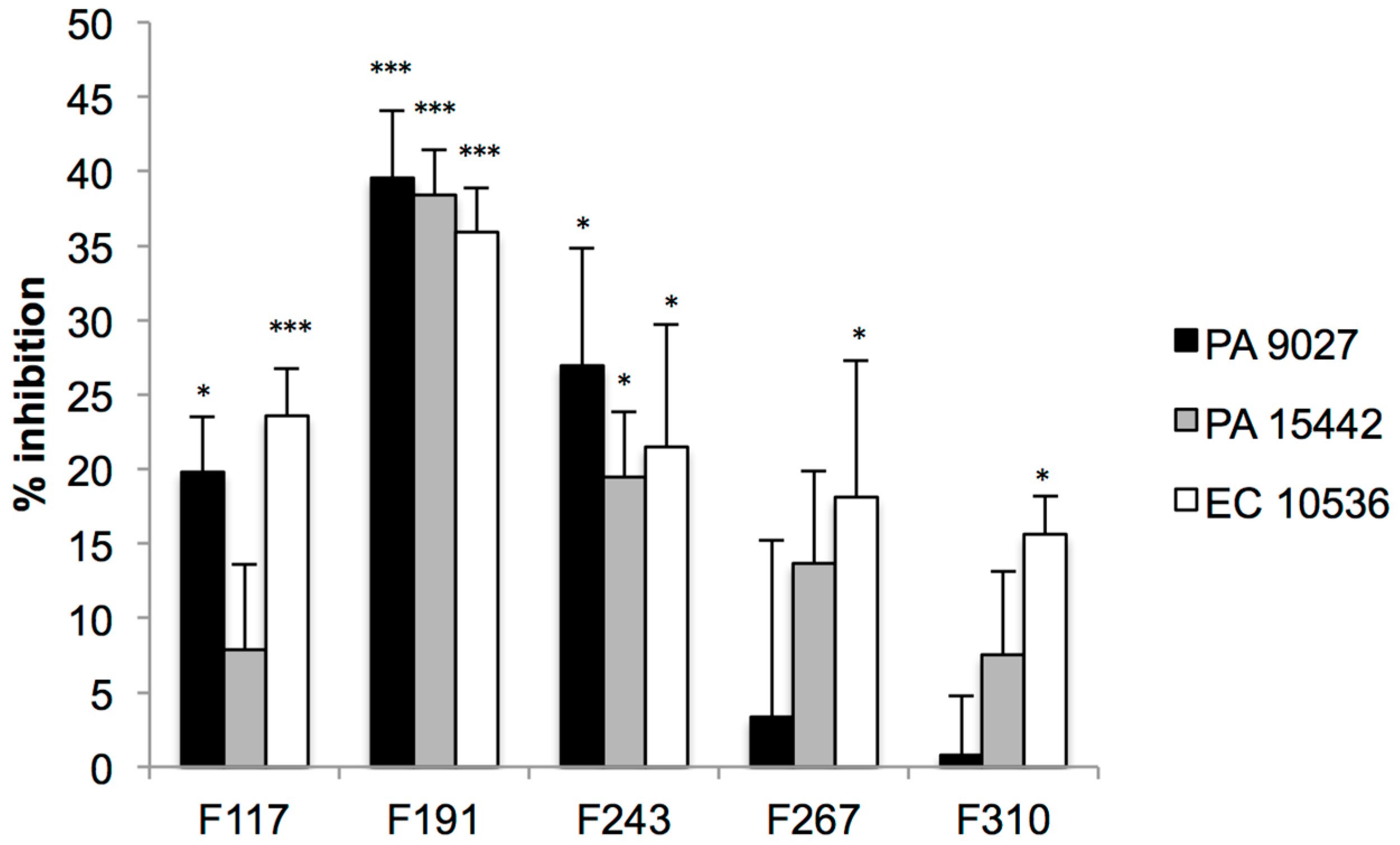
2.2. Screening for QS Inhibition Using a Flavonoid Collection
2.3. Conclusions
3. Materials and Methods
3.1. Bacterial Strains
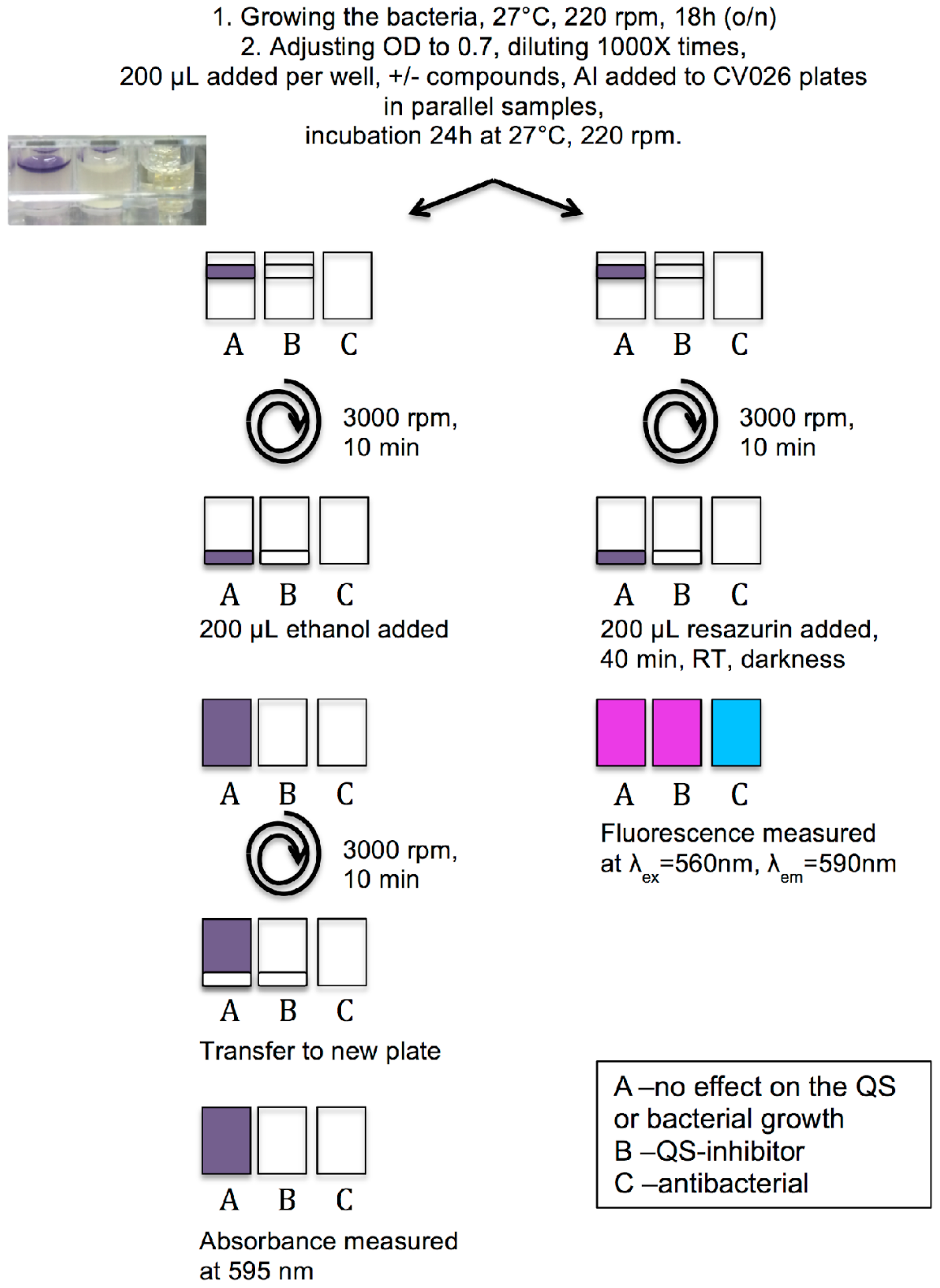
3.2. Optimization of Growing Conditions and Violacein Production by C. violaceum
3.3. Protocol of Violacein Quantification by C. violaceum, Modified and Optimized for 96-Well Plates
3.4. Measurement of Viability of C. violaceum Biofilms Using Resazurin Staining
3.5. QSI Screening of the Flavonoids Collection Using the Optimized C. violaceum Platform
3.6. Effect on the Viability of Biofilms Formed by Pathogenic Strains Using Resazurin Staining
3.7. Effect on the Transition from Microcolonies to Full-Formed Biofilms by Pathogenic Strains
3.8. Statistical Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Taga, M.E.; Bassler, B.L. Chemical communication among bacteria. Proc. Natl. Acad. Sci. USA 2003, 100, 14549–14554. [Google Scholar] [CrossRef] [PubMed]
- Fuqua, C.; Greenberg, E.P. Listening in on bacteria: Acyl-homoserine lactone signalling. Nat. Rev. Mol. Cell Biol. 2002, 3, 685–695. [Google Scholar] [CrossRef] [PubMed]
- McClean, K.H.; Winson, M.K.; Fish, L.; Taylor, A.; Chhabra, S.R.; Camara, M.; Daykin, M.; Lamb, J.H.; Swift, S.; Bycroft, B.W.; et al. Quorum sensing and Chromobacterium violaceum: Exploitation of violacein production and inhibition for the detection of n-acylhomoserine lactones. Microbiology 1997, 143, 3703–3711. [Google Scholar] [CrossRef] [PubMed]
- Fuqua, C.; Greenberg, E.P. Self perception in bacteria: Quorum sensing with acylated homoserine lactones. Curr. Opin. Microbiol. 1998, 1, 183–189. [Google Scholar] [CrossRef]
- Popat, R.; Cornforth, D.M.; McNally, L.; Brown, S.P. Collective sensing and collective responses in quorum-sensing bacteria. J R Soc Interface 2015, 12. [Google Scholar] [CrossRef] [PubMed]
- Donne, J.; Dewilde, S. The challenging world of biofilm physiology. Adv. Microb. Physiol. 2015, 67, 235–292. [Google Scholar] [PubMed]
- Scutera, S.; Zucca, M.; Savoia, D. Novel approaches for the design and discovery of quorum-sensing inhibitors. Expert. Opin. Drug Discov. 2014, 9, 353–366. [Google Scholar] [CrossRef] [PubMed]
- Schuster, M.; Sexton, D.J.; Diggle, S.P.; Greenberg, E.P. Acyl-homoserine lactone quorum sensing: From evolution to application. Annu. Rev. Microbiol. 2013, 67, 43–63. [Google Scholar] [CrossRef] [PubMed]
- Brackman, G.; Coenye, T. Quorum sensing inhibitors as anti-biofilm agents. Curr. Pharm. Des. 2015, 21, 5–11. [Google Scholar] [CrossRef] [PubMed]
- Castillo-Juarez, I.; Maeda, T.; Mandujano-Tinoco, E.A.; Tomas, M.; Perez-Eretza, B.; Garcia-Contreras, S.J.; Wood, T.K.; Garcia-Contreras, R. Role of quorum sensing in bacterial infections. World J. Clin. Cases. 2015, 3, 575–598. [Google Scholar] [PubMed]
- Duran, M.; Faljoni-Alario, A.; Duran, N. Chromobacterium violaceum and its important metabolites-review. Folia. Microbiol. (Praha) 2010, 55, 535–547. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Zheng, H.; Tang, Y.; Yu, W.; Gong, Q. Eugenol inhibits quorum sensing at sub-inhibitory concentrations. Biotechnol. Lett. 2013, 35, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Vasavi, H.S.; Arun, A.B.; Rekha, P.D. Anti-quorum sensing activity of psidium guajava l. Flavonoids against Chromobacterium violaceum and Pseudomonas aeruginosa PAO1. Microbiol. Immunol. 2014, 58, 286–293. [Google Scholar] [CrossRef] [PubMed]
- Vasavi, H.S.; Arun, A.B.; Rekha, P.D. Anti-quorum sensing potential of Adenanthera pavonina. Pharmacognosy Res. 2015, 7, 105–109. [Google Scholar] [PubMed]
- Martinelli, D.; Grossmann, G.; Sequin, U.; Brandl, H.; Bachofen, R. Effects of natural and chemically synthesized furanones on quorum sensing in Chromobacterium violaceum. BMC Microbiol. 2004, 4, 25. [Google Scholar] [CrossRef] [PubMed]
- Taganna, J.C.; Quanico, J.P.; Perono, R.M.; Amor, E.C.; Rivera, W.L. Tannin-rich fraction from terminalia catappa inhibits quorum sensing (QS) in Chromobacterium violaceum and the QS-controlled biofilm maturation and lasa staphylolytic activity in Pseudomonas aeruginosa. J. Ethnopharmacol. 2011, 134, 865–871. [Google Scholar] [CrossRef] [PubMed]
- Linthorne, J.S.; Chang, B.J.; Flematti, G.R.; Ghisalberti, E.L.; Sutton, D.C. A direct pre-screen for marine bacteria producing compounds inhibiting quorum sensing reveals diverse planktonic bacteria that are bioactive. Mar. Biotechnol. (N.Y.) 2015, 17, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Chaudhari, V.; Gosai, H.; Raval, S.; Kothari, V. Effect of certain natural products and organic solvents on quorum sensing in Chromobacterium violaceum. Asian Pac. J. Trop. Med. 2014, 7 Suppl. 1, S204–S211. [Google Scholar] [CrossRef]
- Choo, J.H.; Rukayadi, Y.; Hwang, J.K. Inhibition of bacterial quorum sensing by vanilla extract. Lett. Appl. Microbiol. 2006, 42, 637–641. [Google Scholar] [CrossRef] [PubMed]
- Deryabin, D.G.; Tolmacheva, A.A. Antibacterial and anti-quorum sensing molecular composition derived from quercus cortex (oak bark) extract. Molecules 2015, 20, 17093–17108. [Google Scholar] [CrossRef] [PubMed]
- Cushnie, T.P.; Lamb, A.J. Recent advances in understanding the antibacterial properties of flavonoids. Int. J. Antimicrob. Agents 2011, 38, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Friedman, M. Overview of antibacterial, antitoxin, antiviral, and antifungal activities of tea flavonoids and teas. Mol. Nutr. Food Res. 2007, 51, 116–134. [Google Scholar] [CrossRef] [PubMed]
- Havsteen, B.H. The biochemistry and medical significance of the flavonoids. Pharmacol. Ther. 2002, 96, 67–202. [Google Scholar] [CrossRef]
- Yin, H.; Deng, Y.; Wang, H.; Liu, W.; Zhuang, X.; Chu, W. Tea polyphenols as an antivirulence compound disrupt quorum-sensing regulated pathogenicity of Pseudomonas aeruginosa. Sci. Rep. 2015, 5, 16158. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Huang, X.; Zhang, F.; Feng, L.; Li, J. Inhibition of quorum sensing, biofilm, and spoilage potential in Shewanella baltica by green tea polyphenols. J. Microbiol. 2015, 53, 829–836. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, M.V.; Ortega-Ramirez, L.A.; Gutierrez-Pacheco, M.M.; Bernal-Mercado, A.T.; Rodriguez-Garcia, I.; Gonzalez-Aguilar, G.A.; Ponce, A.; Moreira Mdel, R.; Roura, S.I.; Ayala-Zavala, J.F. Oregano essential oil-pectin edible films as anti-quorum sensing and food antimicrobial agents. Front. Microbiol. 2014, 5, 699. [Google Scholar] [CrossRef] [PubMed]
- Guerin, T.F.; Mondido, M.; McClenn, B.; Peasley, B. Application of resazurin for estimating abundance of contaminant-degrading micro-organisms. Lett. Appl. Microbiol. 2001, 32, 340–345. [Google Scholar] [CrossRef] [PubMed]
- Sandberg, M.E.; Schellmann, D.; Brunhofer, G.; Erker, T.; Busygin, I.; Leino, R.; Vuorela, P.M.; Fallarero, A. Pros and cons of using resazurin staining for quantification of viable Staphylococcus aureus biofilms in a screening assay. J. Microbiol. Methods 2009, 78, 104–106. [Google Scholar] [CrossRef] [PubMed]
- D′Andrea, G. Quercetin: A flavonol with multifaceted therapeutic applications? Fitoterapia 2015, 106, 256–271. [Google Scholar] [CrossRef] [PubMed]
- Vandeputte, O.M.; Kiendrebeogo, M.; Rasamiravaka, T.; Stevigny, C.; Duez, P.; Rajaonson, S.; Diallo, B.; Mol, A.; Baucher, M.; El Jaziri, M. The flavanone naringenin reduces the production of quorum sensing-controlled virulence factors in Pseudomonas aeruginosa PAO1. Microbiology 2011, 157, 2120–2132. [Google Scholar] [CrossRef] [PubMed]
- Gopu, V.; Meena, C.K.; Shetty, P.H. Quercetin influences quorum sensing in food borne bacteria: In-vitro and in-silico evidence. PLoS ONE 2015, 10, e0134684. [Google Scholar] [CrossRef] [PubMed]
- Uroz, S.; Dessaux, Y.; Oger, P. Quorum sensing and quorum quenching: The yin and yang of bacterial communication. Chem. Bio. Chem. 2009, 10, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Eumkeb, G.; Siriwong, S.; Phitaktim, S.; Rojtinnakorn, N.; Sakdarat, S. Synergistic activity and mode of action of flavonoids isolated from smaller galangal and amoxicillin combinations against amoxicillin-resistant Escherichia coli. J. Appl. Microbiol. 2012, 112, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Manner, S.; Skogman, M.; Goeres, D.; Vuorela, P.; Fallarero, A. Systematic exploration of natural and synthetic flavonoids for the inhibition of Staphylococcus aureus biofilms. Int. J. Mol. Sci. 2013, 14, 19434–19451. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Zhang, L. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 2015, 6, 26–41. [Google Scholar] [CrossRef] [PubMed]
- Dekimpe, V.; Deziel, E. Revisiting the quorum-sensing hierarchy in Pseudomonas aeruginosa: The transcriptional regulator RhlR regulates LasR-specific factors. Microbiology 2009, 155, 712–723. [Google Scholar] [CrossRef] [PubMed]
- Walters, M.; Sperandio, V. Quorum sensing in Escherichia coli and salmonella. Int. J. Med. Microbiol. 2006, 296, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Shimada, K.; Ogasawara, H.; Yamada, K.; Shimura, M.; Kori, A.; Shimada, T.; Yamanaka, Y.; Yamamoto, K.; Ishihama, A. Screening of promoter-specific transcription factors: Multiple regulators for the sdiA gene involved in cell division control and quorum sensing. Microbiology 2013, 159, 2501–2512. [Google Scholar] [CrossRef] [PubMed]
- Skogman, M.E.; Vuorela, P.M.; Fallarero, A. Combining biofilm matrix measurements with biomass and viability assays in susceptibility assessments of antimicrobials against Staphylococcus aureus biofilms. J. Antibiot. (Tokyo) 2012, 65, 453–459. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples of the compounds are not available from the authors.

| IC50 μM | 95% Confidence Intervals | IC50 mg·L−1 | |
|---|---|---|---|
| F117 | 21.81 | 18.15 to 26.20 | 7.16 |
| F191 | 19.83 | 17.73 to 22.18 | 5.36 |
| F243 | 8.73 | 6.09 to 12.52 | 2.36 |
| F267 | 3.69 | 2.92 to 4.67 | 1.10 |
| F310 | 23.35 | 19.11 to 28.53 | 7.68 |
| Z′ for the follow up studies: 0.53 ± 0.2 | |||
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Skogman, M.E.; Kanerva, S.; Manner, S.; Vuorela, P.M.; Fallarero, A. Flavones as Quorum Sensing Inhibitors Identified by a Newly Optimized Screening Platform Using Chromobacterium violaceum as Reporter Bacteria. Molecules 2016, 21, 1211. https://doi.org/10.3390/molecules21091211
Skogman ME, Kanerva S, Manner S, Vuorela PM, Fallarero A. Flavones as Quorum Sensing Inhibitors Identified by a Newly Optimized Screening Platform Using Chromobacterium violaceum as Reporter Bacteria. Molecules. 2016; 21(9):1211. https://doi.org/10.3390/molecules21091211
Chicago/Turabian StyleSkogman, Malena E., Sonja Kanerva, Suvi Manner, Pia M. Vuorela, and Adyary Fallarero. 2016. "Flavones as Quorum Sensing Inhibitors Identified by a Newly Optimized Screening Platform Using Chromobacterium violaceum as Reporter Bacteria" Molecules 21, no. 9: 1211. https://doi.org/10.3390/molecules21091211